Chapter 2
The basics of genetics
What you’ll learn in this chapter
In this chapter, you’ll learn the basics about:
- What genes do, and how;
- Why computation and biomedical engineering are important;
- The structure of DNA; and
- How our genetic code might relate to any traits we express.
Warning (or challenge): This chapter is heavy.
It’s meant to be a reference for the rest of the book. So there’s a lot of information here.
You don’t need to understand or remember it all in order to grasp the key ideas about genetic testing.
Feel free to skim this chapter, skip it, come back to it… whatever you like.
If you’re new to understanding genetics or just want a refresher, we do suggest you tackle at least a bit of this material.
If you’ve already done your graduate work in genetics, go ahead and breeze on past.
A quick review of key terms
Before we get into this chapter, let’s learn a few basic terms.
(You’ll also find any bolded term in our glossary.)
DNA is a biological molecule that holds the code for making all living things.
Fun factoid!
Some viruses do not have DNA. Some (like parvovirus) have single-stranded DNA genomes. Some (such as pox viruses) have double-stranded DNA genomes. Most have RNA genomes of some kind.
Genes are regions of DNA that encode instructions for making specific proteins.
If genes have names, we put those in italics, like this: FGF21. This helps us tell them apart from their corresponding protein names.
For instance:
- The FGF21 gene (written in italics) codes for a protein called fibroblast growth factor 21, or FGF21 (no italics).
- The TAS2R38 gene codes for a protein called taste receptor 2 member 38, or TAS2R38.
Variants of the same genes are known as alleles or polymorphisms.
For instance, the type of earwax you have is controlled by a single gene known as ABCC11 (which is also involved, by the way, in how your sweat smells). If you have one of two variants / alleles of the gene, you’ll have wet earwax; if you have a third variant / allele, your earwax will be dry.
Genetics is the study of genes, how they work, and how particular traits (such as eye color) are passed from parent to offspring (known as heredity).
The expression of one gene can influence two or more apparently unrelated processes or traits. We’ll see examples of this as we look at specific topics, such as metabolism. This is known as pleiotropy (from the ancient Greek pleion, or “more”, and tropos, or “way”). A variant in one gene may have a wide variety of effects.
Our genotype is our genetic code; our phenotype is how that code is actually expressed as it interacts with our environment. The same genotype can have different phenotypes. Different environments (for instance, our activity, our nutrition, our exposure to toxins, and so forth) can change our observable traits (for instance, our physiology or behavior).
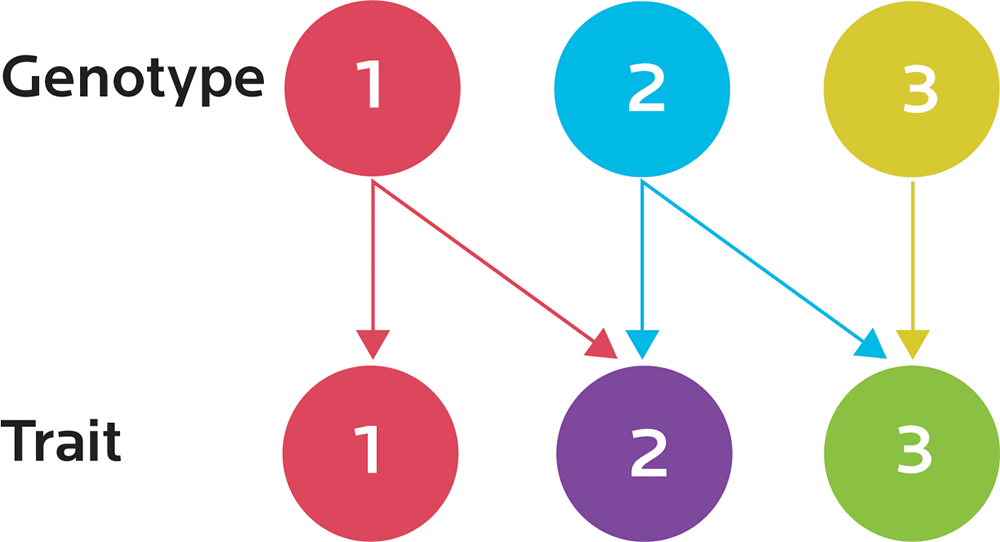
Epigenetics is the study of how activation and inhibition of gene expression is regulated. The same genetic “blueprint” may be used to express different things; for instance, exercising can activate expression of genes involved in the antioxidant response. We may already have had those genes, but exercising affects their epigenetic expression.
A genome is the complete set of an organism’s genetic information. For instance, the Human Genome Project studied the complete genetic code of human beings.
In this book, we’ll be talking about genetic testing.
Generally, this doesn’t mean we are testing the entire genome. As you’ll learn, the amount of information in an entire human genome is very, very, very large. We have about 22,000 genes, and about 3 billion base pairs, or pairs of nucleotides in our strings of DNA. (We’ll learn more about nucleotides in a moment.)
With genetic testing, we usually test a single gene or single nucleotide polymorphism (SNP), which is just one single point mutation to one nucleotide within a gene. There can be several SNPs in the same gene; most widely used SNPs are in noncoding regions.
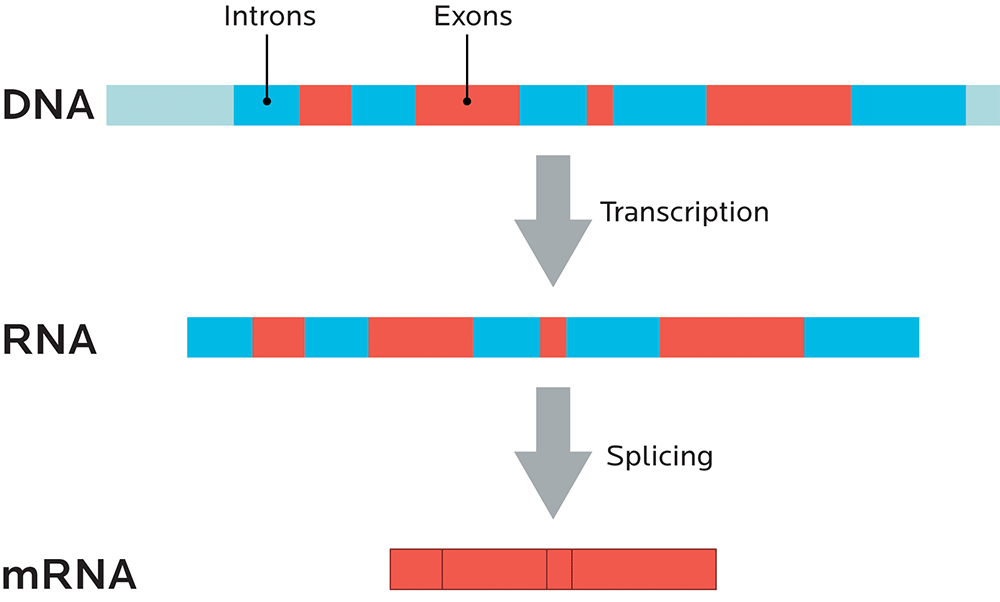
Noncoding regions are parts of the genetic code that don’t code for any proteins, and they’re known as introns. Regions that do code for proteins are known as exons. (We’ll look more at coding and noncoding regions in Chapter 4, and at splicing more below.)
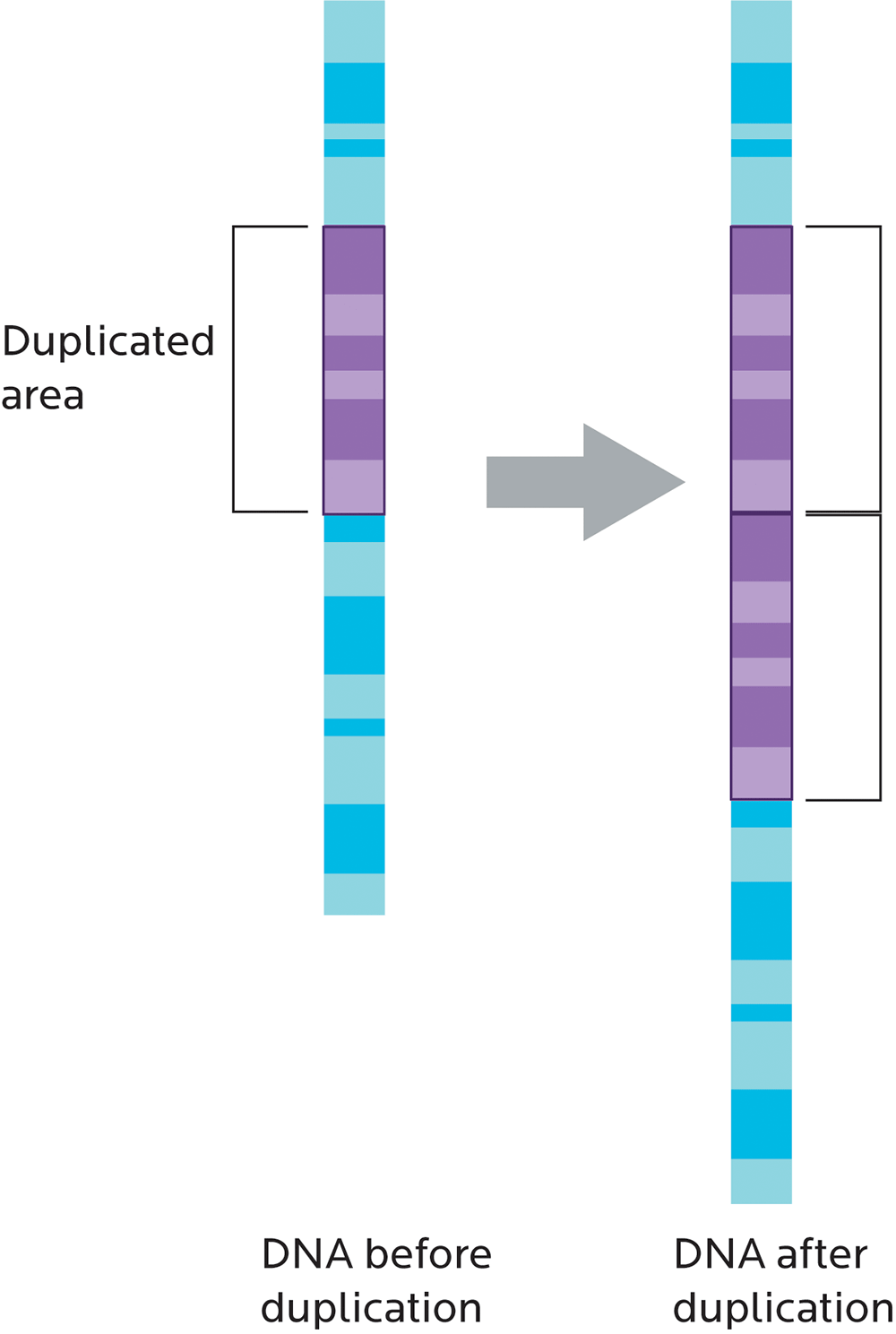
Copy number variation (CNV) refers to whether chunks of genetic material are repeated, kind of like having three or more of the same socks instead of a single pair.
CNVs can also affect how genes work. For instance, having multiple copies of genes that make the enzyme amylase would increase our ability to break down the carbohydrate amylose. This variation may reflect our ancestral history (for instance, whether we come from an ethnic group that has traditionally eaten a high-starch diet) and may be linked to our body weight.
What do genes do?
Genes are information.
In the real world, we have to figure out a few things when we use information:
- How to transmit and transport information — how to get it from one place to another without losing it along the way.
- How to reproduce information properly and carefully.
- How to store information so it doesn’t degrade or take up too much space.
- How we receive and interpret informational signals that are sent.
Genes and their associated products and processes do all of this inside our bodies.
We can think of a gene as a unit of information or a set of instructions for making something — in this case, a specific protein.
In eukaryotic cells (cells that have a nucleus and distinct organelles bound by a membrane), DNA is organized in chromosomes in the nucleus.
When cells divide, chromosomes are duplicated and DNA is replicated. Once cells finish dividing, each cell ends up with its own full set of chromosomes. Eukaryotes store most of their DNA inside the cell nucleus and some of their DNA in organelles, such as mitochondria or chloroplasts (in plants).
Genetically speaking, we are about half of each of our parents. But we’re not exactly like our mom and dad. And we’re not a precise 50% of each. We’re more like a 49-point-something percentage of each, plus some random mutations that sneaked in along the way.
We’ll learn more about mutations in a bit.
For now, the key points are:
- Genes store information.
- Information is passed from parent to offspring.
- It’s not a perfect transmission every time.
When we look at our genes, or particular variation in our genes, we can see this transmission of information. And we can see the variations among ourselves, even within the same family.
- Why does this happen?
- How does this work?
- And what does this all mean for your own genetic code?
To explain, let’s start with a concept that may be a little new to you.
Biology is computation.
Most people tend to think of biology as just a bunch of wet squishy bits. We can also think about biology as being a type of computation.
Imagine you were trying to program a computer to do something.
Computers are pretty literal and only do what you tell them, so you want to make sure you’re absolutely clear. You’ll also need to give the computer some structured parameters for making decisions, otherwise they get confused.
For example, you might tell the computer:
IF this is true, THEN do this thing.
IF today’s date is July 18, THEN send Dr. John Berardi a birthday card.
IF Dr. JB likes cake, THEN make him some.
Maybe your IF / THEN instructions have conditions:
IF Dr. JB likes cake OR you like cake, THEN make a cake.
IF you are on a desert island AND today is July 18, THEN spell out “Happy birthday” using driftwood instead.
Biology works in similar ways.
Let’s imagine something simple, like the communication between a cell and the outside world.
Some cells have receptors (such as a specific type of protein) on their membranes. These receptors bind to other things, such as other proteins or simpler molecules.
Each receptor is picky and will only bind to particular things (these specific molecules are known as ligands), much like a key will only work in one or two locks.
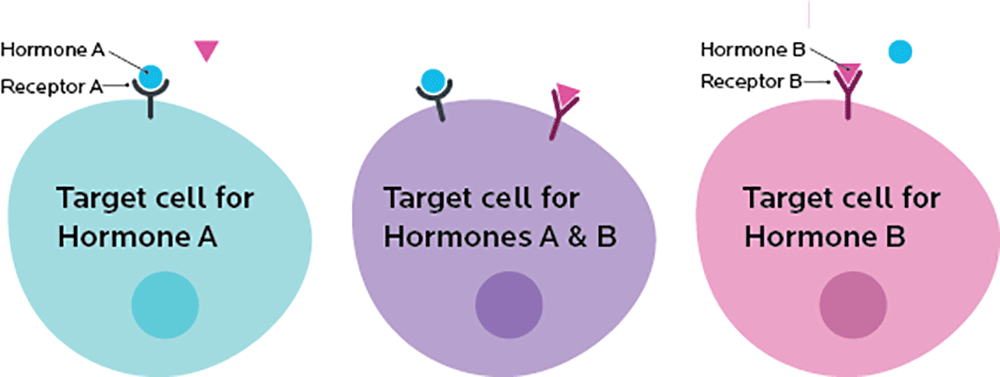
A receptor acts like a switch. If something binds to a receptor, it triggers another event, like turning a switch to “on”.
So, we might imagine a situation where the instructions for our cell’s “biological computer” read something like this:
IF receptor A is “on” AND receptor B is “off”, THEN make protein from gene A
IF gene A is active, AND gene B is available, THEN make the protein from gene B.
Note that this looks very much like the computer code you might have learned in school:
IF xyz THEN do something.
Computers are still pretty dumb, relative to biology. But at a basic level, the concept is similar.
Genes encode a series of instructions, like a computer program… and accumulate problems, also like computer programs.
Computers can start from scratch. In theory, you could make a computer that thinks in a way that nothing ever has before, using components that nobody has ever used before.
Biological systems don’t work that way. They can only use the structures and systems that they have inherited (along with, of course, any new mutations, which are only a small part of a much bigger whole).
Sometimes this can mean that biological systems are pretty efficient — perhaps evolution has done a lot of work to tidy things up.
Sometimes the systems can be less efficient. They have a lot of what computer folks call “cruft”: clutter and redundant junk such as old equipment or code that just hangs around and either does nothing or actively gums up the works.
Biology is engineering.
Remember high school chemistry with diagrams like this?
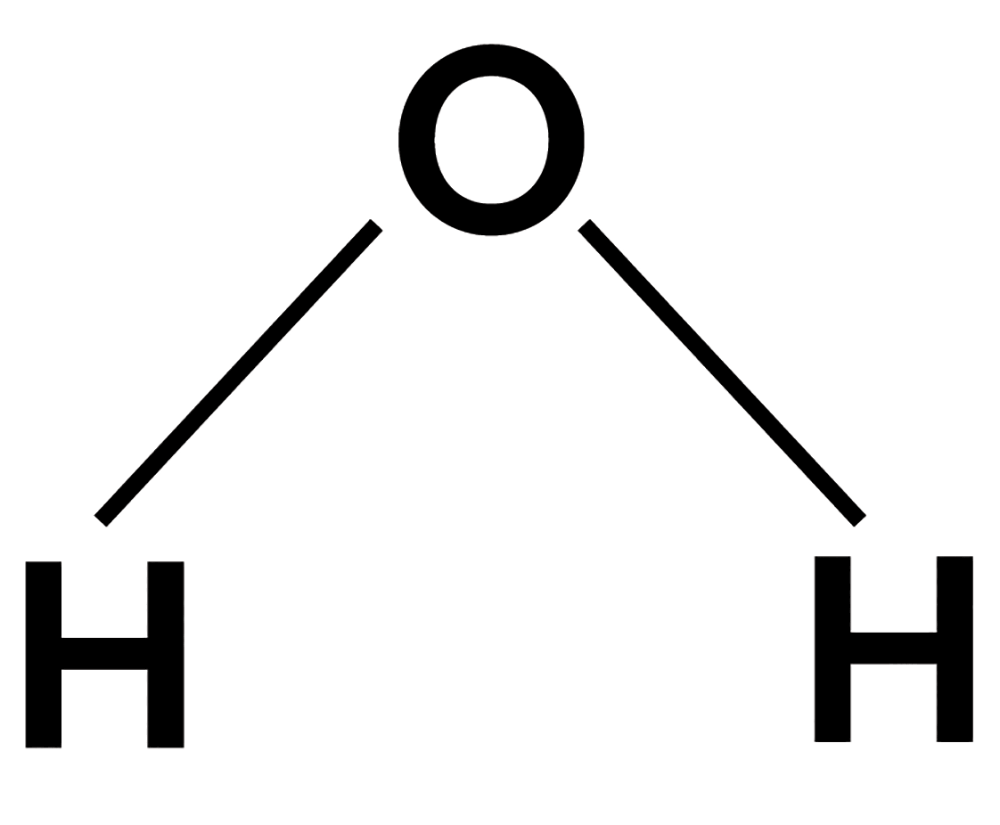
(That’s water.)
Or these?
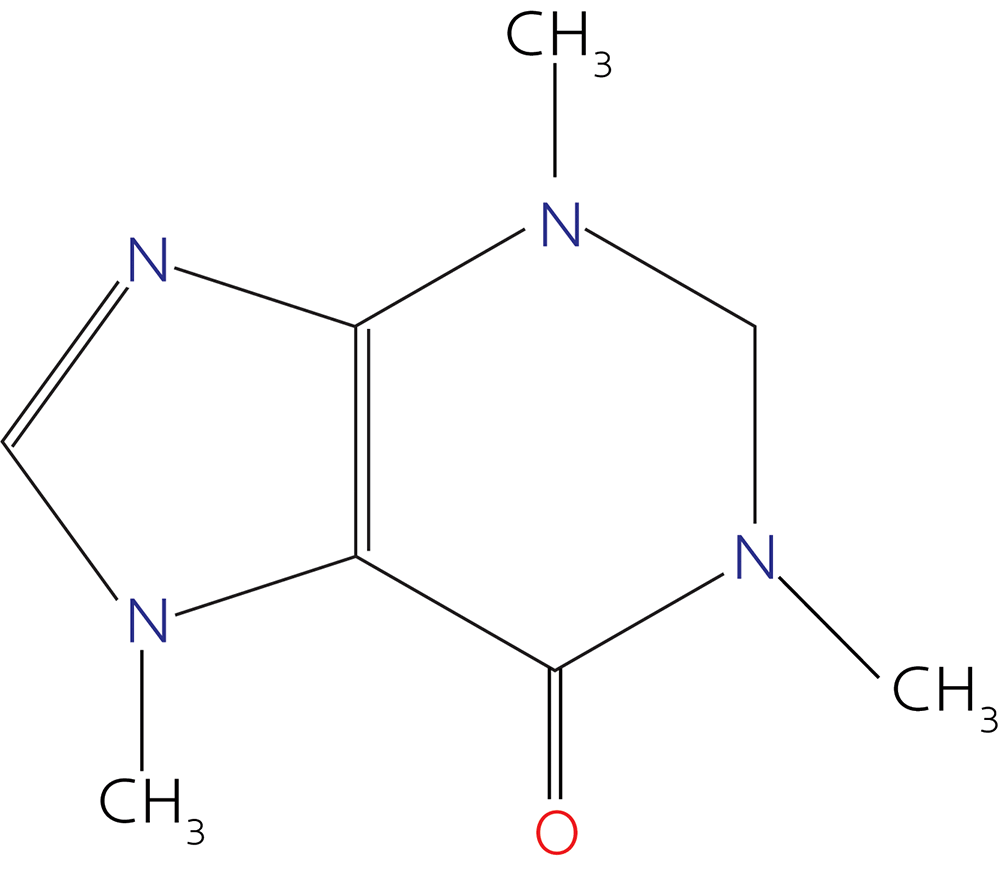
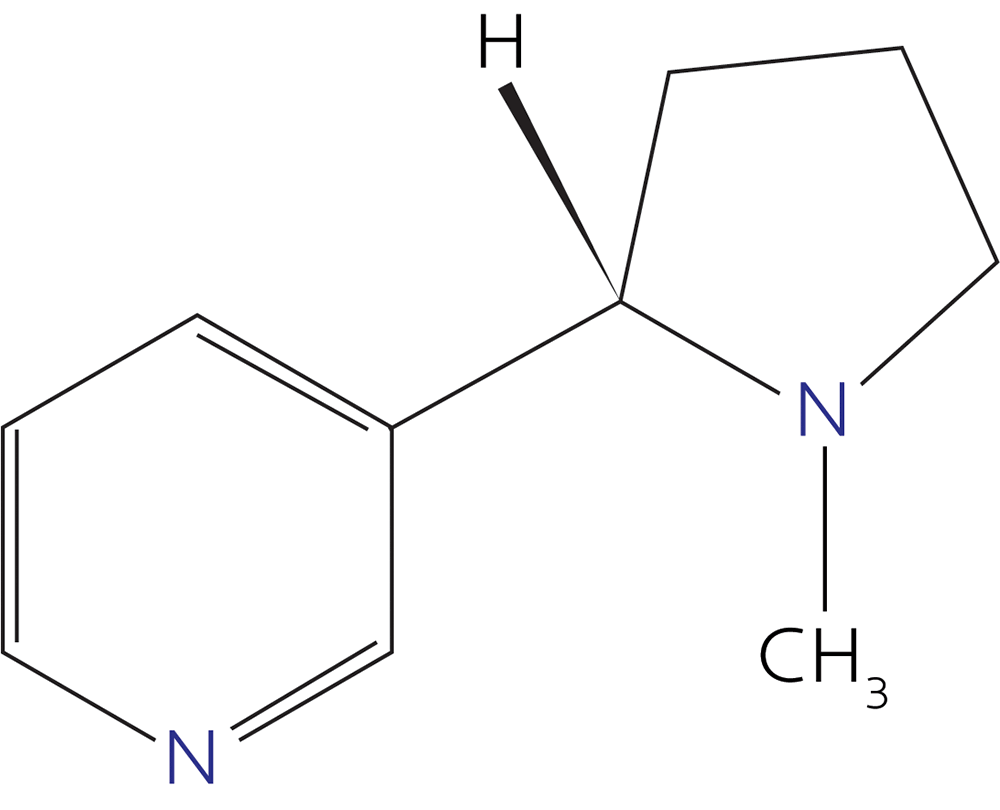
(Those are caffeine and nicotine, maybe two of your best friends in high school, you rebel you.)
You might have been left with the impression that “molecules” are basically flat polygons and lines.
However, molecules are three-dimensional.
So water actually looks more like this:
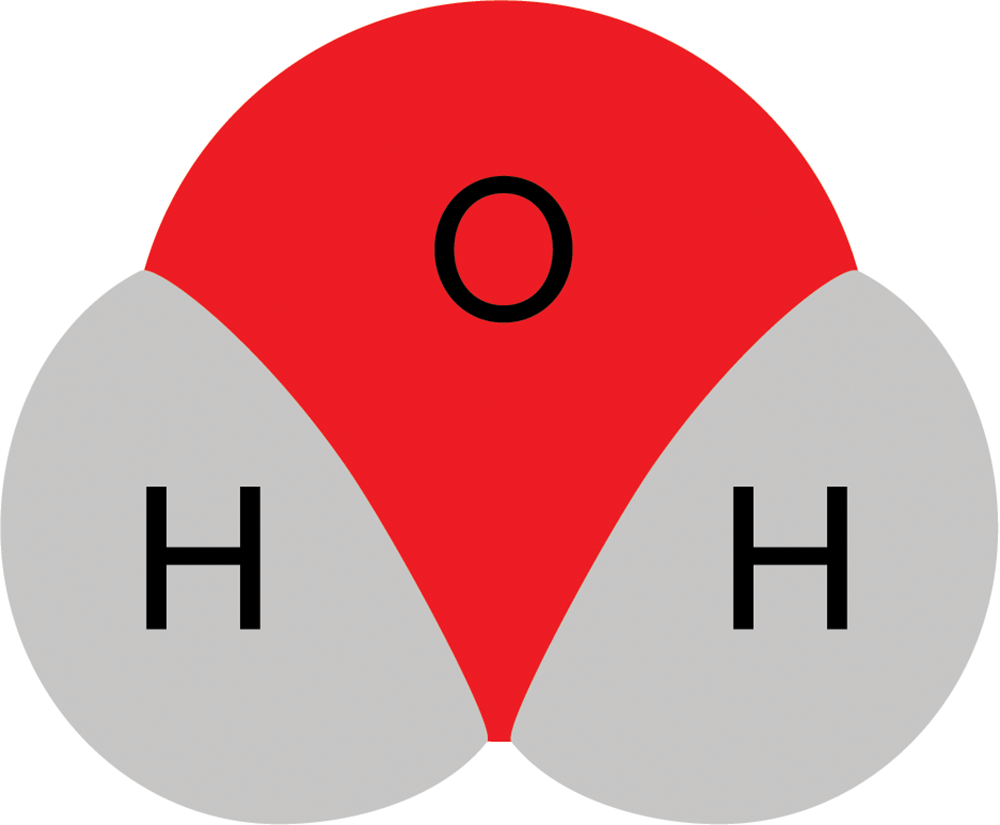
And nicotine looks more like this:
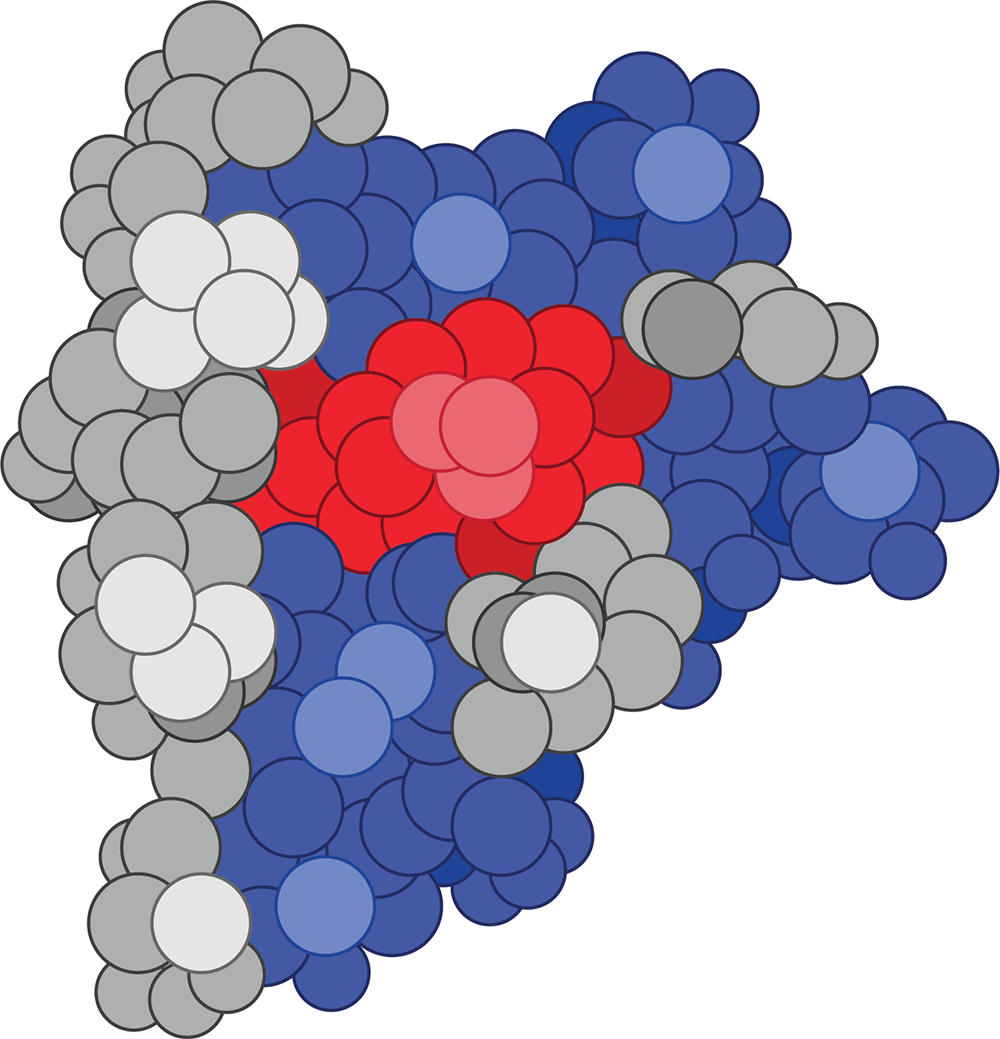
Molecular structures in the body are the same way.
For instance, here is a three-dimensional protein (a Cas9 nuclease, in case you’re wondering) that our co-author Alaina printed in her lab:

You’ll notice that proteins are not abstract line drawings, but actual, tangible objects.
If you blew that nicotine protein up larger, it would look like this:
The physical shape of the protein would affect how you could interact with it. The same thing happens at the molecular level.
For instance, let’s imagine you have a protein that is shaped like this:
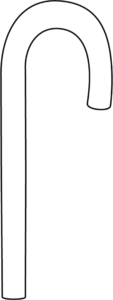
It has a straight end and a hooked end.
Let’s imagine that 3-dimensional J-shaped protein is moving around in your body.
Then it finds a protein that looks like this:
What happens? Well, if conditions are right, maybe the hook-shaped part of the first protein will latch on to the eye-shaped part of the other protein, and stick there.
The physical shapes of the proteins determine how they will interact.
If you just have this:

…then the J-hook has nothing to connect to.
In other words: Shape matters. The physical characteristics of molecules determine whether and how they will interact (or react) with other molecules. And biology is, after all, just a whole lot of reactions with the final result of making something be alive.
DNA is a 3-dimensional structure.
First: What does the acronym “DNA” stand for?
Yes, eager student in the front with your hand up, the answer is deoxyribonucleic acid.
Aside from making you the team champ on trivia night, knowing the full name of DNA actually gives you clues to how DNA is constructed, and to understanding how genetics work. Part of that has to do with the physical shape of DNA and how the molecules fit together.
A DNA strand is shaped like a ladder, and made out of two pieces:
- A deoxyribose “backbone”, i.e., the side rails of the DNA ladder. (RNA has ribose here.)
- Bases or nucleotides, i.e., the ladder’s rungs.
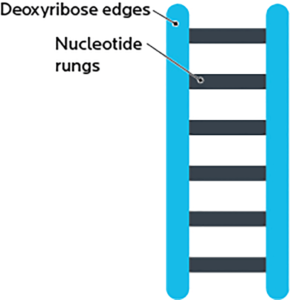
Ribose is a simple sugar. “De-oxy” means “without oxygen”, so “deoxyribose” means a ribose without an oxygen.
OK, now imagine that the deoxyribose sugar is actually a sort-of T-shaped piece of Lego, like this:

And imagine a few of those Legos stacked on top of each other, like this:

Then imagine you’re getting real fancy with the Lego stack and you want to indicate which way is up.
So you draw an arrow along the stack, like this.

Congratulations. You just built the deoxyribose backbone of DNA.
And, since you have 3 pokey-outy parts to which other things can attach, you have also built the foundation for a specific unit of DNA, known as a codon (aka a sequence of three nucleotides).
DNA uses computation.
Except imagine that in DNA computation, we aren’t making numbers, but amino acids.
Remember our Lego structure? We can stick bases, or nucleotides, on it. Bases are the building blocks of nucleic acids.

With DNA, we have 4 options for nucleotides:
- Adenine (A)
- Cytosine (C)
- Guanine (G)
- Thymine (T)
RNA (which we’ll look at in a moment) gives us one more: uracil (U), which replaces thymine.
Two of these nucleotides (A and G) are long. Two (C and T) are short. So in a full strand of DNA, short nucleotides pair with long ones, and vice versa.
Like this:
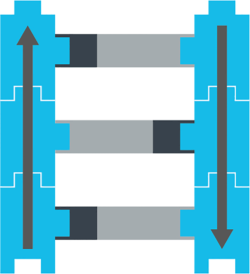
The nucleotide formula for proteins
You can imagine that each nucleotide is like a box that could have 1 of 4 possible nucleic acids inside: either an A, a C, a G, or a T, like this:

With 2 boxes, we get 4 x 4 values = 16 possible combinations.
With 3 boxes, we get 4 x 4 x 4 values = 64 possible combinations.
DNA codes for proteins. Proteins are made of amino acids. So part of that DNA has to code for a particular amino acid.
We need to make about 20 amino acids.
We can’t do that with 1 box (which can only contain 1 of 4 possible nucleotides), or 2 boxes (which only gives us 16 possible combinations). We need 3 boxes.
Hence, a codon is 3 units. BIO MATH!!
We need many different 3-nucleotide codons to code for these 20 amino acids. Keeping it all straight can be confusing. Thus, researchers have come up with some ways to help us catalogue and interpret codons, such as a codon wheel (shown below).
A codon wheel helps us in both directions: We can decipher what amino acid a particular codon codes for, and what codons code for a particular amino acid.
To read the wheel, start from the center. That’s one nucleotide. Then go outwards and pick another one of 4. And outwards again, picking another one of 4.
For instance:
- Starting with C in the middle → moving outwards and picking another C → then moving outwards again and picking a U, C, A or G gives you the amino acid leucine.
- Starting with G in the middle → moving outwards and picking an A → then moving outwards again and picking an A or a G gives you glutamine.
You might also notice that some amino acids can be made with more than one combo of nucleotides.
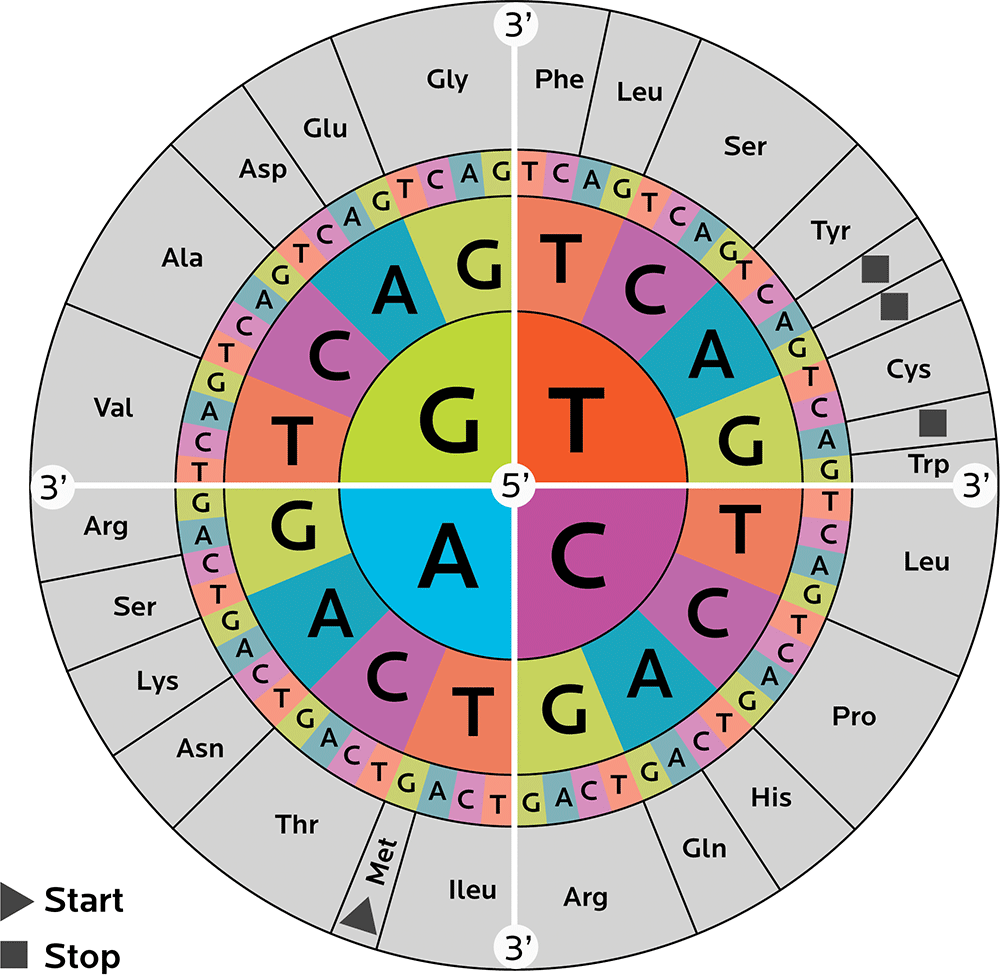
We’ll learn more about codons and how they work later.
For now, just remember this structure of DNA:
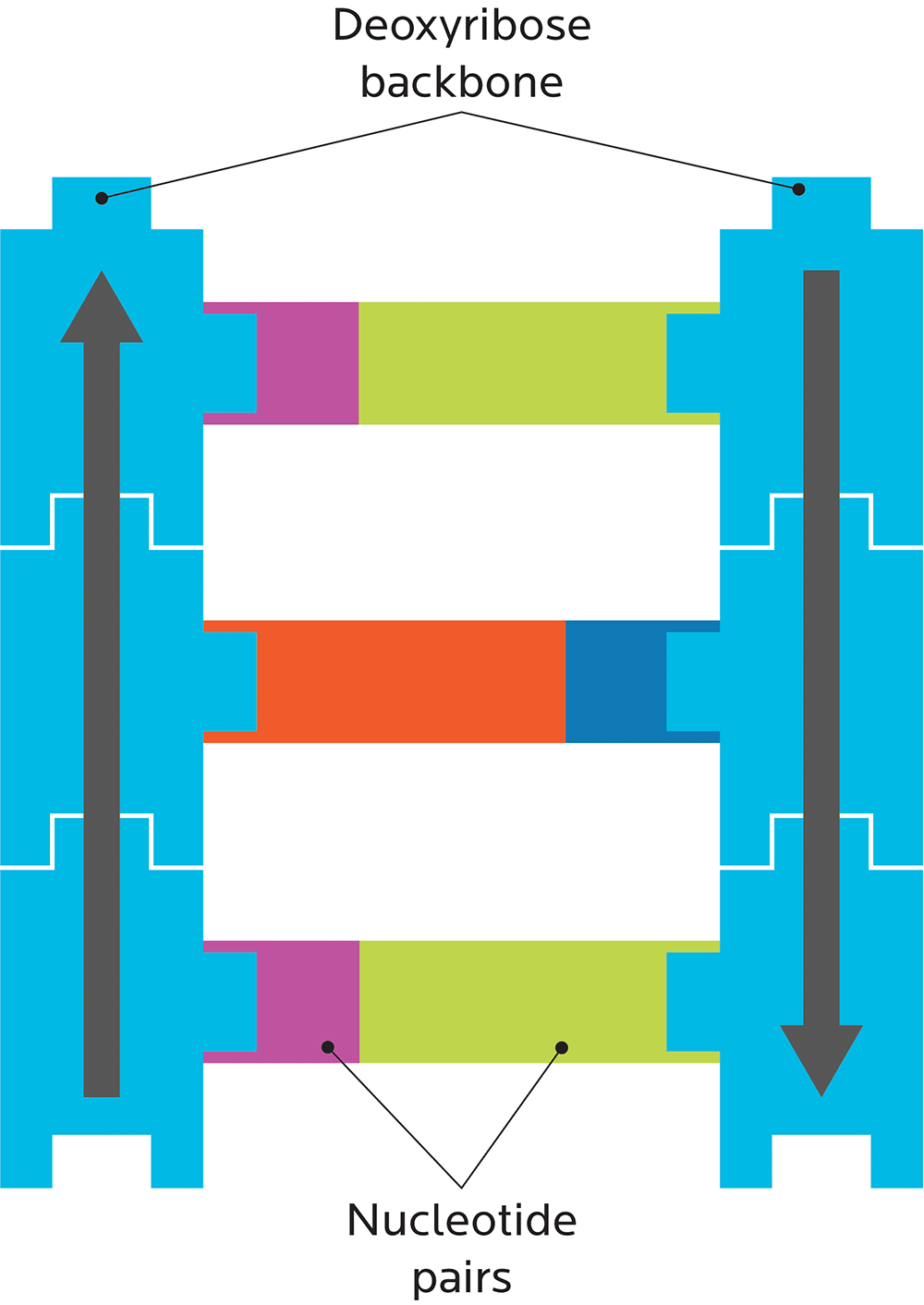
Genes have “geography”.
The shape of DNA is not random.
To go back to the idea that genes store information, this shape is not an accident. Because of the way the DNA Lego blocks fit together:
- The molecule is extremely stable.
- Our bodies can copy it with extremely high fidelity and accuracy.
This is good, because as we’ll see later, we don’t really want things falling apart or getting sloppy with reproduction. Errors happen all the time, but we have biological proofreaders to catch them.
Remember our little ribose Lego? Deoxyribose is a carbohydrate, which means it has carbon. In biochem, we number the carbons. Each little Lego block has one carbon called the 5-prime (written as 5’) and another called the 3-prime (written as 3’).
Our bodies will always read DNA in a certain order (hence the arrow): from 5-prime to 3-prime, and they go in opposite directions. One side will be 5’ to 3’ reading up, the other 5’ to 3’ reading down.
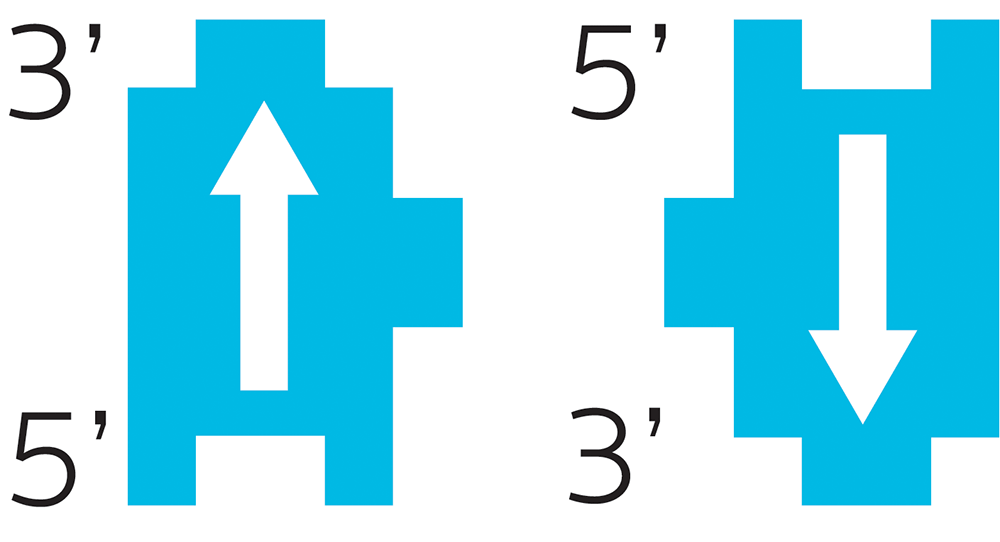
This directionality of each DNA piece is important.
Think about how you read. If you read English:
YOU READ FROM LEFT TO RIGHT.
.TFEL OT THGIR MORF DAER T’NOD UOY
DNA reading works the same way. It has to go in a particular direction.
Imagine you were making a machine to read DNA. It’s a simple computer, so it can only read one nucleotide at a time. It scans down the strand, reading from 5’ to 3’, reading the name of each nucleotide, like this:
A C T T G A A T G C A T C
and so on. It has to go in that direction, because the reverse (reading from 3’ to 5’) would be something totally different:
C T A C G T A A G T T C A
Different “letters”, different meaning.
DNA isn’t just floating around randomly in cells. It’s packed into highly organized structures, tightly wrapped around proteins called histones like beads on a string, so that massive amounts of this genetic material can fit into the tiny space of a cell’s nucleus.
Unspooled, your strands of DNA would be around two meters long. Yet they fit into a space about 1/1,000 of a millimeter.
This protein-DNA complex is called chromatin, and it’s what makes up your body’s chromosomes.
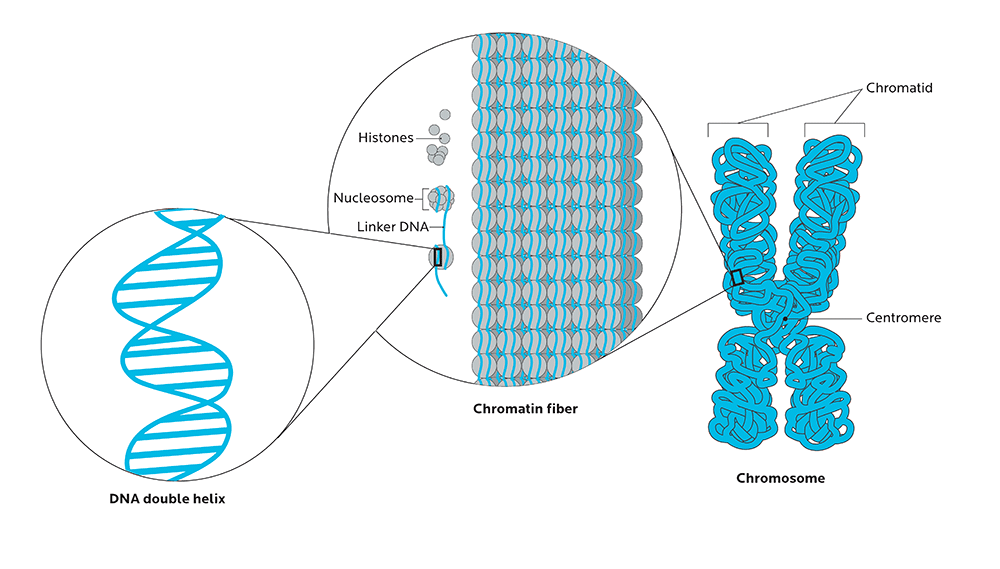
The ways that our chromosomes are folded in space, and physically organized in the nucleus, can significantly affect how genes are expressed.
For example:
- Factors known as enhancers, which increase the likelihood that a given gene will be transcribed, need to be located close to the regions that they act on.
- Much of epigenetics (which we’ll look at briefly below), or the regulation of gene expression, is related to histone modification, or changes to the physical configuration of the histone proteins that DNA wraps around.
Recent groundbreaking research at the Salk Institute for Biological Studies has just revealed that we are now able to see how chromatin is organized in a living cell. (Previously, we had to pull the cell apart to see this, so we could not see how chromatin worked “in the wild”.)
This research revealed that the way in which chromatin was packed into the nucleus — its shape as well as its density — affected its function.
What this means is that:
- The shape of molecules is important. Shape affects how molecules work, and how they interact with other molecules.
- Thus, genetic expression is not just about the genes we have, but where they’re located.
- Commercial genetic testing can tell us about some of the specific genes we have, but not about how they’re physically organized. We can’t know the geography of our genes from a commercial test, which means we can’t know a lot about how those genes may be expressed.
How are proteins made from genes?
The central dogma of molecular biology
All of this adds up to what is sometimes known as the central dogma of molecular biology: DNA to RNA to proteins.
- DNA is read in groups of three nucleotides called codons.
- These DNA codons are transcribed to an “in-between” molecule, messenger ribonucleic acid (mRNA). (We’ll look at this in more detail in a minute.)
- The mRNA is read and each mRNA codon is translated to one amino acid.
- Amino acids are linked together to make proteins.
Thus, again, DNA’s job is to store the information to make proteins.
All of molecular biology revolves around this basic principle.
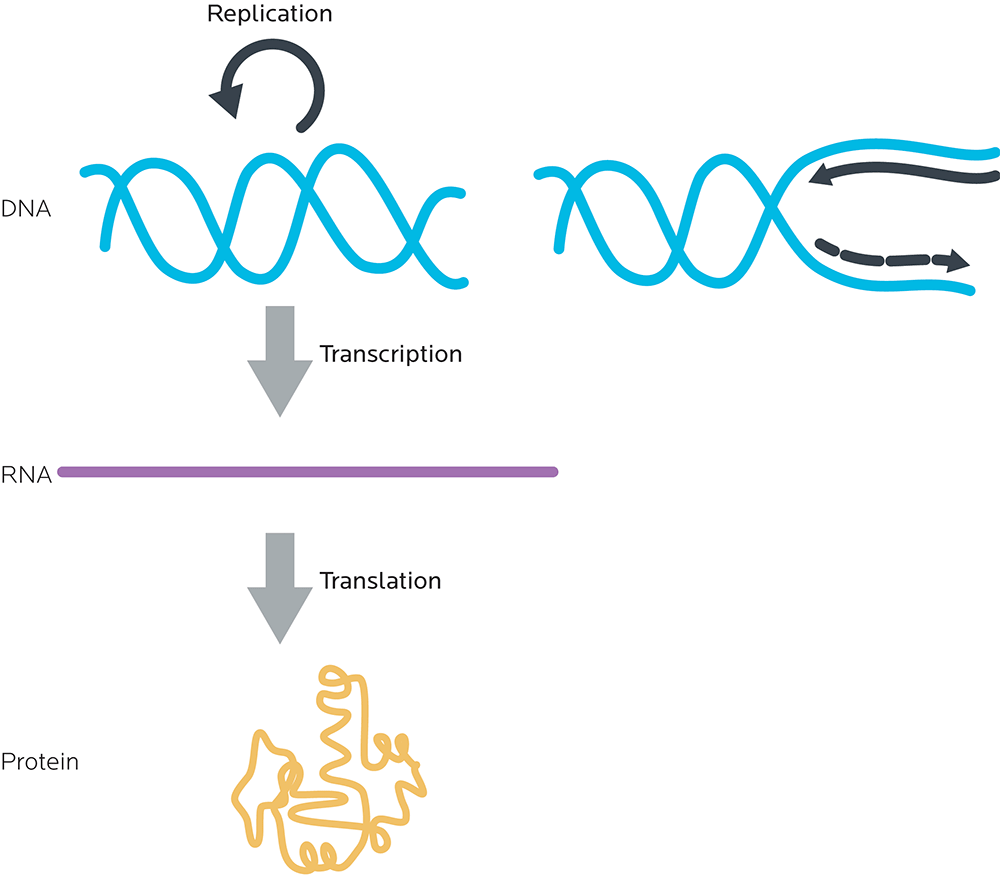
Gene expression and making proteins
There are two general steps to make a protein. Together, they’re called gene expression.
- RNA synthesis (transcription): Making RNA, using the instructions from DNA. At this step, double-stranded DNA is “unzipped” like a zipper by enzymes called polymerases into single, complementary strands of RNA.
- Protein synthesis (translation): Making proteins, using the instructions from RNA.
RNA has many jobs, and there are several types, which we’ll look at in a moment.
For now, we’ll focus on messenger RNA (mRNA). mRNA is the intermediary between DNA and proteins.
For a long time, scientists thought that one gene coded for one mRNA, that then coded for one protein: If you had 100 genes, you’d make 100 different mRNAs, and then 100 different proteins. That way, if you knew all the genes, you’d know all the proteins.
It’s more complicated than that. (Warning: We’re going to say this a lot. Like, a lot.)
RNA splicing
One gene can code for many related proteins through RNA splicing.
RNA splicing is like producing alternative cuts of movies — think of director’s cuts, alternative endings, 20 year anniversary edition cut, and what-have-you-done-to-my-favorite-movie?? cut.
Some cuts could be minor. Some could completely change the movie (i.e., the protein that’s expressed).
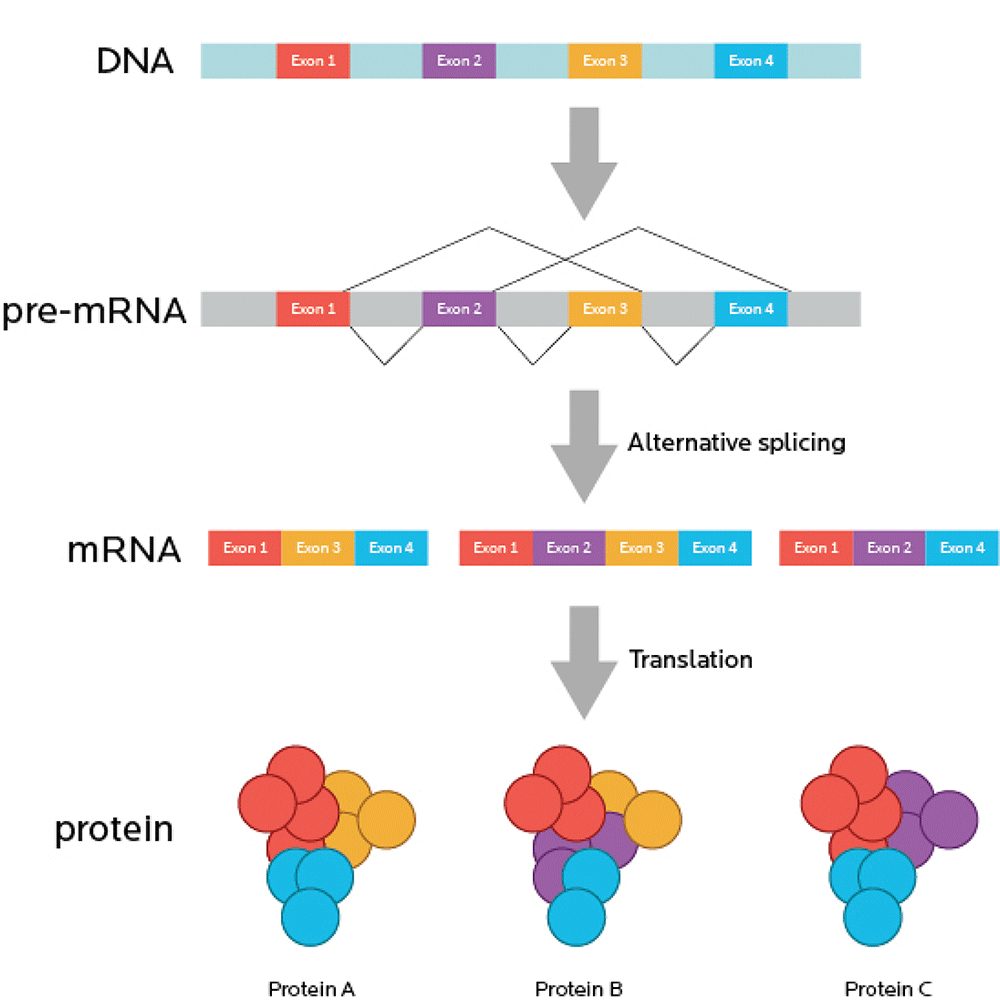
Now, imagine your movie is also full of commercials. You have to watch the movie in little bits. This is annoying. So, imagine that you re-cut the movie, take out the commercials, and stick all the movie bits back together so you get one uninterrupted full-length feature.
You can think of introns (sometimes known as “intervening sequences”) as the commercials. Remember, introns don’t code for proteins.
You can think of exons (sometimes known as “expressing sequences”) as the movie itself. Since exons code for proteins, they’re like the “story” of what will happen, genetically speaking.
Just like a movie with irritating and useless commercials removed, mRNA is spliced so that all the introns are taken out, and only the exons, the expressing sequences, remain.
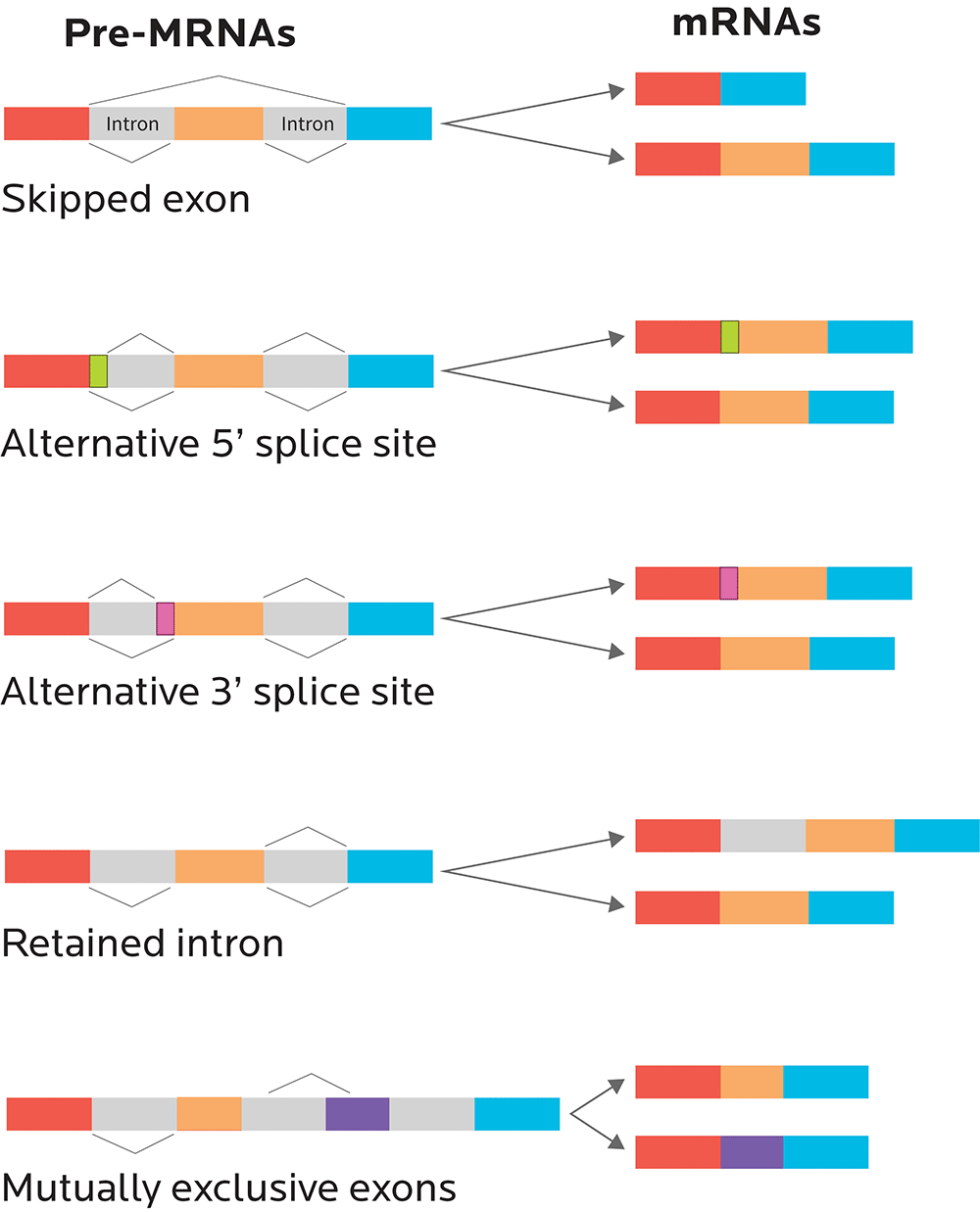
Now, sometimes there’s a small problem: Our editor is excellent but not perfect.
So, when our “mRNA movie” is “re-cut” during the RNA splicing process:
- We might end up with a perfect, commercial-free copy of the original movie. Just some nice exons all stuck together in an uninterrupted, faithfully reproduced sequence.
- Or, if alternative splicing occurs, we might end up with a few scenes missing from our movie. A few exons might get cut out.
Transporting mRNA in and out of the nucleus
Remember that DNA and RNA live mainly in the nucleus. (In Chapter 6, we’ll talk about another kind of DNA, mitochondrial DNA, that doesn’t.)
Before it can be used to code for protein, mRNA has to clear a couple more hurdles. Enter mRNA export.
- mRNA needs to get out of the nucleus and to ribosomes, the protein-making factories in the cytoplasm.
- It also needs to keep itself from breaking down. If it’s broken down (degraded), then it can’t be used.
You don’t want RNA just randomly escaping the nucleus, just like you don’t want drunken texts to your ex or boss escaping your phone at 3:00 AM.
Thus, you need to regulate mRNA degradation and stability so it can make it to the ribosome and be translated into a protein.
Nuclear exporting involves nuclear transport receptors that chaperone the mRNA through a nuclear pore, which means that trying to get the mRNA out of the nucleus can be a bit of a bottleneck.
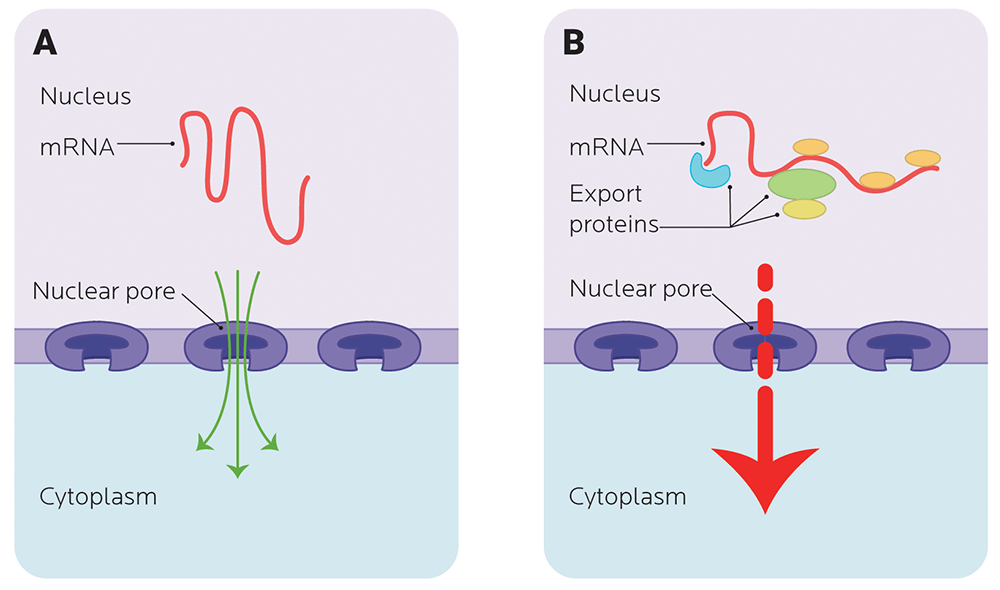
Once precursor mRNA grows up and becomes mature mRNA (via splicing and some accessories stuck on for stability), it’s selected and moved out of the nucleus.
Mature mRNA has a bunch of extra nucleotides stuck on one end (a 3’ poly-A tail) that acts a bit like the little plastic ends on your shoelaces that keep them from getting frayed. On the other end of the mRNA (the 5’ end), it has a cap (a 7-methylguanosine cap).
The cap and the tail protect the mRNA from being broken down and keep it stable. The more stable the mRNA is, the more protein it can make.
Other types of RNA
Besides pre-mRNA and mRNA, there are several other types of RNA we need to make proteins.
- Transfer RNA (tRNA). tRNAs are RNA molecules that carry specific amino acids to the ribosome. They match mRNA codons with their respective amino acid.
- Ribosomal RNA (rRNA) is an RNA enzyme that links amino acids together in the ribosome to make a polypeptide chain. Remember that ribosomes are where the mRNA is read and matched with tRNA that carry amino acids, which are then assembled to make the protein.
Other RNAs — such as small nuclear RNA (snRNA), microRNA (miRNA), and small interfering RNA (siRNA) — regulate how much and what type protein will be made.
There’s no exam.
Just remember:
It’s much more complicated than “One gene, one protein.”
| Type of RNA | Main function |
|---|---|
| mRNA | Codes for protein |
| rRNA | An RNA enzyme that links amino acids together |
| tRNA | Bridges between mRNA codons and the amino acids they code for |
| snRNA | RNA splicing |
| miRNA | Generally block translation of specific mRNA, though can also upregulate (increase) it |
| siRNA | Selectively target specific mRNA for degradation |
Controlling gene expression: from DNA to protein
There are 5 steps going from DNA to protein that are controlled via mRNA production, processing and transport.
- Transcription: making RNA from DNA
- RNA processing: splicing RNA
- RNA export: exporting mRNA out of the nucleus
- Translation: making protein from mRNA
- Post-translational processing: getting rid of unwanted or harmful mRNA, which can happen before proteins are made (kind of like a quality control mechanism)
Your body doesn’t constantly make RNA from DNA. It’s a very regulated process. Only certain genes are transcribed, and only at certain times.
Genes have three control regions:
- promoters
- enhancers
- terminators
Promoters and terminators (basically starters and stoppers) are similar for all genes.
But enhancer regions are different. Their variations allow them to match transcription factors with some specificity, allowing systems to activate and deactivate transcription of certain genes by the presence and absence of their transcription factor(s).
How do mutations happen?
Think about nucleotide pairs like matching pairs of socks. Ideally, you have all your socks properly sorted and paired after doing laundry. Sometimes, that happens. Sometimes, not.
Sometimes you lose or mismatch socks — maybe one sock, maybe a pair of socks, maybe a few pairs. Sometimes you even end up with new socks… socks that aren’t even yours.
The same can happen with the nucleotide pairs of DNA. This is called a mutation — some type of permanent change to the nucleotide pairing sequence. This can happen in a few different ways, and for different reasons.
The example of antioxidants and reactive oxygen species
You may have heard the term “antioxidant” (for instance, as an ingredient in “superfoods”, or vitamins C and E).
But why do we need antioxidants in the first place?
In normal metabolism, we create what are called reactive oxygen species (ROS). ROS are unstable, oxygen-containing molecules that are normal byproducts of cellular respiration. Oxidative damage happens naturally during cell metabolism — some estimates suggest this occurs 10,000 times a day per cell.
Usually, our cells are able to balance oxidative damage with their own antioxidant system.
However, under environmental stress (such as exposure to toxins), ROS can build up faster than the cell can clear them. This can cause mutations by chemically modifying the nucleotides to become a slightly different molecule.
Types and causes of mutations
Having a lot of ROS hanging around is one cause of mutations.
There are many other ways for oxidative changes to occur, such as:
- exposure to ionizing radiation (such as X-rays or radioactive material);
- certain types of chemicals (such as heavy metals or pesticides);
- ultraviolet light (two nucleotides — cytosine and thymine — are particularly sensitive to UV light); and
- spontaneous (and uncorrected) errors in the DNA replication/copying process.
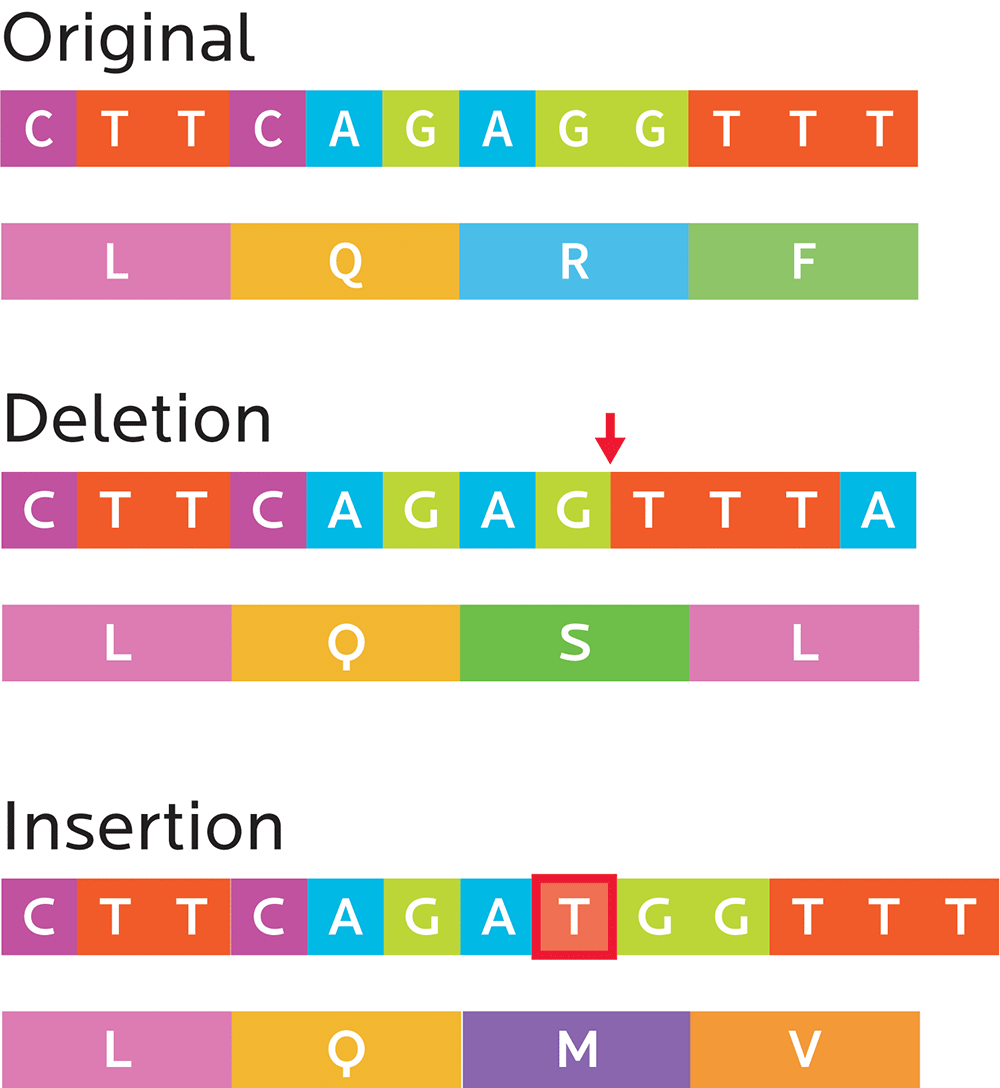
Mutations can affect anything from only a single nucleotide, to large-scale alterations to chromosomes. They can change the structure and function of genes.
Small mutations can sometimes have big effects that change a protein’s amino acid composition:
- Substitution mutations: In a codon, one nucleotide is exchanged for another (for instance, an A for a G or a C
for a T). These can end up being:- Silent mutations that code for the same or a similar-enough amino acid (e.g., both CCA and CCT result in proline)
- Missense mutations, which code for a different amino acid (e.g., CCA results in proline, but CTA results in leucine).
- Nonsense mutations, which create stop codons and can truncate the protein (e.g., TAC makes tyrosine, while TAA signals “stop”). This means that a cell’s ribosome (the protein-making factory), will stop producing the protein before that protein has all of its amino acids. Consequently, this can affect the way the protein works.
- Frameshift mutations: Sometimes, the DNA copying process can insert or delete one or more nucleotides, which means that the whole reading frame has shifted. Everything after the frame might now be nonsense, or the gene may simply be missing some amino acids.
- Insertions, as the name implies, add one or more extra nucleotides into the DNA.
- Deletions remove one or more nucleotides from the DNA.
More significant mutations that can affect one or more entire genes can include:
- Amplifications or duplications, or more than one copy of genes or regions of a chromosome.
- Deletions of large parts of a chromosome, which results in losing some genes in those regions.
- Bringing together previously separated chunks of DNA, which may mean different genes come together to form new ones (known as fusion genes). An example of this is the unique seaweed-digesting bacterial enzyme in the gut bacteria of people with Japanese ancestry, which researchers speculate may originally have come from the genes of marine bacteria that live on seaweed. This “jump” of DNA from unicellular organisms (such as bacteria) to multicellular organisms (such as humans) is known as horizontal gene transfer.
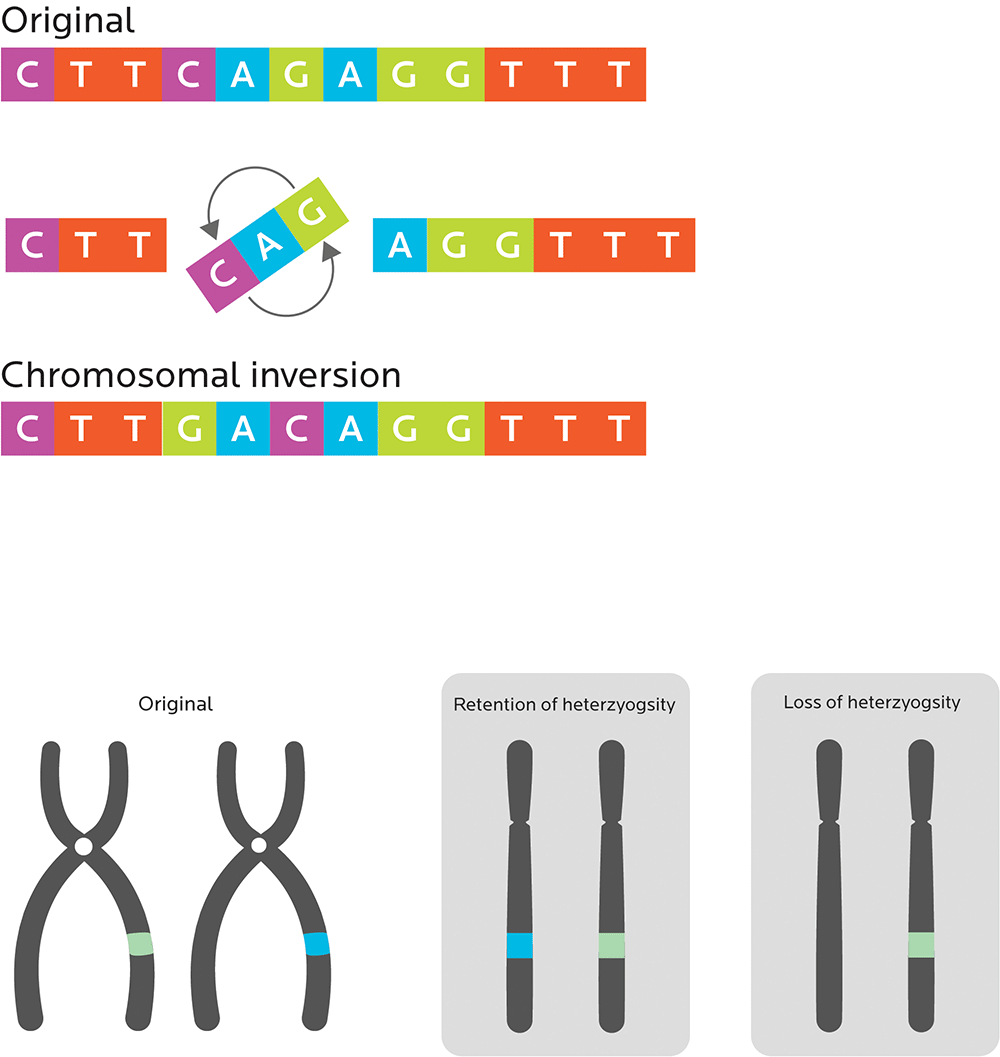
- Chromosomal translocations, which happen when unrelated chromosomes swap parts.
- Terminal and interstitial deletions, which happen when a piece of DNA is removed from a single chromosome. Large deletions can be severe or even catastrophic to an organism. Prader-Willi Syndrome, which affects many aspects of normal growth, development, and metabolism, is a result of an interstitial deletion.
- Chromosomal inversions, which “flip” the order of a segment of a chromosome. These mutations are usually benign.
- Loss of heterozygosity, or losing an allele from the (possibly different) pair of genes that you inherit from your parents, so that you only get one “regular” allele.
Sometimes mutations have observable effects, sometimes not.
For instance:
- Genes can be partly or completely “de-activated”. These are known as loss-of-function mutations, or inactivating mutations. For example, in androgen insensitivity syndrome (AIS), a mutation inactivates the androgen receptor and can cause chromosomally XY people to develop physically as female.
- The effects of genes can get stronger. If we have extra copies of a gene in a genome (copy number variation, or CNV), there are more copies of the gene to be transcribed, and the effect can be more pronounced.
- Genes can change their function. For example, a mutation at codon 6 of HBB, the β-globin gene (which changes the amino acid code from glutamic acid into valine) can mean that hemoglobin can’t carry as much oxygen. You may know of this mutation as sickle-cell anemia.
- Sometimes mutations are helpful, perhaps giving us some kind of advantage in our environment. Sometimes they are harmful. Sometimes they have no apparent effect at all.
- Mutations that gave us an advantage in one environment (such as the ability to store energy when food was scarce and required lots of effort to get) might not give us an advantage in a different environment (e.g., now that food is abundant, cheap, and energy-dense).
However, sometimes mutations kill an organism. These are known as lethal mutations. While there are some exceptions, if you’ve made it to adulthood, you’ve likely escaped most of the major lethal mutations.
Fixing DNA damage
How does the body reach that high fidelity we mentioned earlier? By precisely spotting and quickly repairing some of the more common mutations.
Like the Quality Assurance process of factory production, our bodies carefully check for DNA damage and quality at various “checkpoints” in the cell cycle.
If the “QA testers” spot damage, they (ideally) stop the “production line”, pause cell division, and fix things.
In fact, given how many components our genome has, it’s quite amazing how effective and efficient our genetic copying process is — as accurate as only one major error per every 1010 (or 10,000,000,000) nucleotides. Try finding a factory with that level of precision!
Our bodies can repair DNA damage in several ways:
- Mismatch repair (MMR) happens when a set of genes “notices” errors in DNA replication and recombination.
- We know of 7 DNA MMR proteins (MLH1, MLH3, MSH2, MSH3, MSH6, PMS1 and PMS2) that work in an orderly process to find and fix things.
- When MMR doesn’t happen correctly, we may get what is called microsatellite instability (MSI), which means that small, variable, highly mutation-prone chunks of DNA (aka microsatellites) end up as part of our genetic material. MSI has been linked to many cancers.
- Base excision repair (BER) and nucleotide excision repair (NER) processes fix lesions and physical damage that can lead to DNA mis-pairing or breaks during replication. Deficiencies in BER / NER have been linked to cancer as well as neurodegeneration.
- Nonhomologous end-joining (NHEJ) and homologous recombination repair (HRR) are processes that repair double-strand lesions, or breaks. You can think of double-strand breaks as your DNA “ladder” being cut between the rails. These are some of the most dangerous types of DNA damage. One double-strand break is enough to kill a cell or destroy the integrity of its genome.
- Translesion synthesis (TLS) DNA polymerases let unrepaired lesions get through the process of DNA replication, to be fixed later.
Sometimes, critical mutations occur not to particular genes that do things like make body structures, but to the genes involved in finding and fixing mistakes. So diseases of genetic origin occur from not having a strong enough “repair crew”.
You could spend your life studying the details of genetics. (Heck, we had a hard time keeping this “basics” chapter under 10,000 words.)
But you don’t need to know all the details if you simply want to grasp the basics of genetic testing.
Just get the general idea:
Mutations and variations are complex.
Sometimes mutations take an organism out of the game before it can reproduce. Or, if the organism does manage to reproduce, the mutation isn’t passed to its offspring.
Sometimes mutations don’t affect the reproduction process (for instance, it’s something that only affects people over 60, or it gives you a funny-shaped nose but someone loves you anyway).
Often, mutations and genetic variations can be inherited, and as a result, they can affect our health, nutrition, and fitness.
Early in genetic research, scientists often wondered if a single gene could be responsible for particular diseases. For instance, could there be a “cancer gene”?
We now know that most diseases result from combinations of factors.
For instance, the BRCA1 and BRCA2 gene variants play a strong role in the development of breast and ovarian cancers. Many other genes do too, such as genes involved in DNA repair.
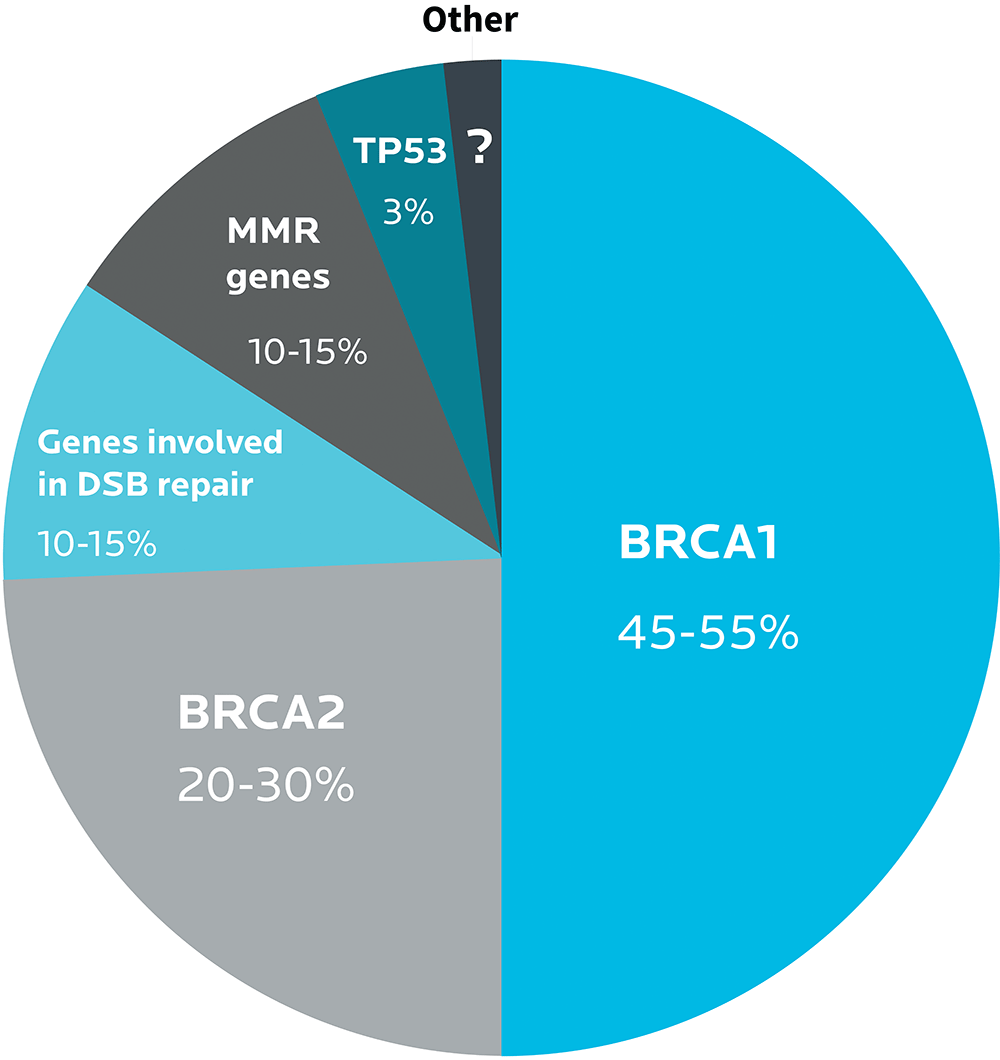
Different types of ovarian cancer
| Type 1 | Type 2 | |
|---|---|---|
| Mutations | PTEN, KRAS, BRAF, PIK3CA, ERBB2, CTNNB1, ARID1A, PPP2R1A, and microsatellite instability | TP53 BRCA1 BRCA2 |
| Prevalence | About 30% | About 70% |
| Tumor type | Serous, endometrioid, mucinous, and clear-cell tumors | Serous, mixed malignant mesodermal tumors carcinosarcomas, and undifferentiated tumors |
| Grade & progression | Low and borderline, slow and often isolated to ovary | High and aggressive |
These variations affect not only the origins of particular types of ovarian cancer, but also their locations, clinical progressions and outcomes.
Diseases such as cancer are not just one “thing”.
They are diverse phenomena that result from complex genetic and environmental interactions.
We’ll be emphasizing this throughout the text:
As with most preferences, health risks, and genetic traits, there are many complex, interrelated factors.
There is almost never one single gene that inevitably leads to a given result.
Any genetic data we share are simply clues for further exploration.
Epigenetics: Environment matters.
Have you ever met identical twins that were different? Different personalities, different habits, maybe even different physical traits — despite being genetically the same?
Studies have compared what happens when one identical twin exercises or eats nutritiously and the other one doesn’t. For example, one study compared 10 pairs of twins. One twin of each pair exercised regularly; the other twin was sedentary.
The researchers found that the more active twins were leaner and metabolically healthier. They also had more grey matter in their brains than their genetically identical but inactive counterparts. Other studies have found similar results.
Why aren’t two people with the same genetic blueprint exact copies of one another?
The answer is epigenetics, the regulation of whether or not our genes are expressed.
Our environment (such as what we eat, what’s around us, our exercise habits, what happened to us as children, and so forth) affects gene expression.
There are many ways that this can happen, such as histone modification. You’ll remember that histones are proteins that make up part of the package of DNA, and can affect how parts of that DNA are activated or repressed.
A full discussion of epigenetics is cool, but beyond the scope of this book.
Just get the general ideas here:
- Genetic expression depends on more than just the genetic blueprint we received at conception.
- Just knowing our genetic code (our genotype) is not enough to fully understand what it means, or how it may shape our phenotype (for instance, our health, our risk of disease, our physical characteristics like height, and so forth).
- Many other factors can affect how and whether specific genes are expressed.
What’s up next
Now that you have some fundamentals under your belt, we’ll explore how genetic tests work, and what they look for.





