Chapter 9
What we found: Food intolerances
What you’ll learn in this chapter
In this chapter, we’ll look at:
- the basic physiology of immune system and inflammatory responses;
- how food allergies, intolerances, and sensitivities work;
- how these can be affected by our genes; and
- what you can learn from genetic testing about your reactions to particular foods.
In previous chapters, we’ve explored the idea of whether there’s a single “best diet”. By now, it should be obvious there isn’t.
Two important points to keep in mind as you read through:
- While science is cool, and we have some interesting genetic findings and areas for further exploration, we still know comparatively very little.
- Just because a genetic test can tell you about your risk of particular food sensitivities doesn’t mean that it can tell you the “perfect” diet for you.
And remember our usual caution:
As with most preferences, health risks, and genetic traits, there are many complex, interrelated factors.
There is almost never one single gene that inevitably leads to a given result.
Any genetic data we share are simply clues for further exploration.
Almost all of us have eaten something that “didn’t agree with us” in some way.
Of course, this doesn’t mean we have any special sensitivity.
It might just mean we should have said “no” before the third pound of suicide-spice chicken wings.
Aside from situations in which We Really Should Have Known Better, many people find that certain, normally innocuous foods — such as bananas, avocados, berries, shrimp, eggs, etc. — are just bad news for them in general.
In general, we might loosely call these “food sensitivities” or “food intolerances”, but we want to be careful with our language.
It’s important to understand the difference between a food allergy, a food sensitivity, a food intolerance, and a specific disease, such as celiac disease or Crohn’s disease, all of which have a genetic basis.
It’s also important to understand that these and related digestive and autoimmune disorders, like most chronic diseases, are polygenic (i.e. many genes contribute) and emerge from complex interactions between genes, behaviors, and environment.
Antibodies, immunoglobulins, and inflammation
An antibody is a type of protein that is produced by the immune system when it identifies a foreign substance, known as an antigen. Antibodies help identify pathogens (such as bacteria or viruses) as well as allergens and toxins.
Antibodies can also be known as immunoglobulins, and get the abbreviation “Ig”. There are 5 main antibodies: IgA, IgD, IgE, IgG, and IgM.
Antibodies are Y-shaped proteins that share the same general structure, but their tips vary quite a lot. This helps them match a specific antigen.
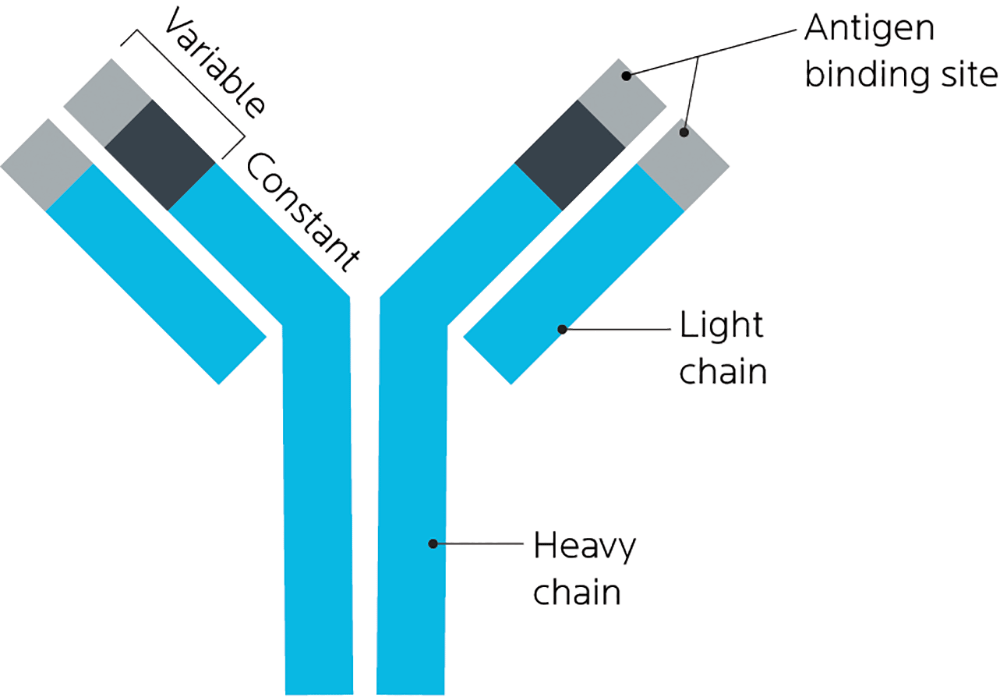
If our bodies think a particular food or part of a food is harmful or foreign, it’ll create a targeted antibody to defend against it.
Over time, with repeated exposure, these immunoglobulins can build up and create a physiological response. This response can be acute (immediate, sudden, often dramatic) or it can be chronic (ongoing, persistent, often lower-grade).
Typically, the response includes some form of inflammation.
Inflammation can be local — restricted to a small area, such as a patch of skin. Or it can be systemic — affecting various parts of our physiology, such as our respiratory system, our joints, our nervous system, and/or our digestion.
We can often see inflammation emerge in real time, with redness, swelling, rashes, hives or a combination of these symptoms. Chemically speaking, we can also “see” inflammation by the presence of particular substances, such as interleukins (IL), histamine, prostaglandins, and so forth.
The expression of all of these, of course, is shaped by our genes.
There are many types of immune and inflammatory responses
Because many types of food-related immune and inflammatory responses can have similar symptoms (such as stomach pain or diarrhea), it can be hard to tell which is which. In addition, people can have more than one health condition (for instance, a food allergy plus celiac disease).
Food allergy
An allergen is something that causes a histamine response, known as an immunoglobulin E (IgE) immune reaction. White blood cells (mast cells and basophils) release histamine molecules when exposed to an allergen and cause an inflammatory response such as:
- hives;
- swelling;
- trouble breathing; and
- a sudden drop in blood pressure.
Allergies are generally characterized by “sudden and strong”. IgE spikes quickly, but also tapers off relatively quickly, generally dissipating within a few days (though it can last up to a week or two).
Food allergies, like other allergies, do seem to run in families. This suggests that we can, to some degree, inherit our allergy risk.
Food sensitivity
Food sensitivities tend to present more with things like abdominal pain and bloating, which are related to the actions of immunoglobulin G (IgG) rather than IgE, as in allergies, above. IgG responses tend to be slower than IgE responses, often taking hours or even days to show up.
People with health problems such as inflammatory bowel syndrome / disorder (IBS / IBD), Crohn’s disease, or ulcerative colitis tend to have higher IgG levels than healthy controls. IgG antibodies can also infiltrate and affect other tissues, such as the pancreas, thyroid, respiratory system, kidney, lymph nodes, or salivary glands.
Food intolerance
Other types of food intolerances, such as the example of lactose intolerance, below, are typically caused by not making enough of the right types of enzymes (for instance, lactase). Without enough of a particular enzyme, we can’t digest some foods properly.
Autoimmune-related problems
Gluten sensitivity and celiac disease
Gluten is a protein found in wheat, barley, and rye. It’s a type of storage protein known as prolamins, and made up of two proteins: gliadin and glutenin. (Barley prolamins are hordeins, rye prolamins are called secalins and oat prolamins are avenins).
Gluten is what gives wheat its elasticity and viscosity so that it can be made into bread, pasta, or other baked goods.
You may have heard of “gluten intolerance” or even “gluten allergy”. You may have heard that gluten is the root of all human disorder and dysfunction. (Move over, money and power! Gluten is the new gangster in town!)
As with all things, humans are diverse.
Some people — those blessed with iron constitutions — seem to be able to eat anything. They munch on the bread basket cheerfully, crumbs falling out of their mouths, apparently unaffected.
Other people notice some sensitivity to gluten and similar proteins in other grains. Perhaps their joints hurt a little bit; perhaps they get a bit of a stuffy nose; perhaps a slight skin rash.
A few people notice a strong, immediate reaction, even to trace amounts of gluten or the proteins in other grains.
Like other food sensitivities, sensitivity to gluten and other grain proteins can come in a few different flavors, ranging from mild to severe.
Why?
Well, at least part of this is, of course, genetic. But we don’t yet know all the factors involved.
Celiac disease
Celiac disease is an autoimmune condition triggered by gluten. Because it’s autoimmune, which means that the body’s immune system attacks its own healthy tissues, symptoms can be widespread through the entire system.
As we’ve seen in our chapter on metabolism and thyroid autoimmunity, genes related to autoimmunity can play many roles. You’ll see here that many autoimmune disorders share common genetic components and there is no single “autoimmunity gene”.
As with most diseases, our risk for celiac disease appears to be polygenic. To date, research suggests that genetic factors can explain about 55% of celiac disease cases.
Not surprisingly, research has found links between celiac disease and several genes related to immune and inflammatory responses, including:
- CCR3 codes for the C-C chemokine receptor type 3. Chemokines are a type of cytokine, or cell signaling molecule, that tell other cells to move somewhere, like directing immune cells to move to the site of an infection. The CCR3 protein is highly expressed in immune system cells such as eosinophils and basophils (types of white blood cells), or T-helper cells, as well as in the epithelial cells of our airway.
- HLA-DQ, which we’ll look at more below.
- IL12A, which codes for interleukin 12A (interleukins are a family of cytokines). IL12A helps to direct the activities of T-helper cells.
- IL18RAP, which codes for interleukin 18 receptor accessory protein. It’s involved in the binding and signaling of IL-18. Variations in this gene have been linked to inflammatory bowel and Crohn’s diseases, as well as leprosy and atopic dermatitis.
- MYO9B, which codes for myosin IXB and is involved in maintaining the integrity of the intestinal lining. It has also been associated with inflammatory bowel conditions. People with variants of MYO9B may have more intestinal permeability, aka “leaky gut”.
- PFKFB3, which codes for a protein that plays a role in cancer progression, circadian clocks, autophagy, and insulin signaling.
- PRKCQ codes for protein kinase C, which is involved in T-cell immune system signaling.
- PTPRK codes for protein tyrosine phosphatase, receptor type K, involved in cell growth, differentiation, migration, and division. Levels of this protein are associated with some types of cancer.
- RGS1 codes for regulator of G-protein signaling 1, which has been used as a marker of intestinal tissue quality in studies of colorectal cancer. It’s also been linked to mental health and multiple sclerosis (along with IL12A).
- SH2B3: Remember this little guy from Chapter 6 on metabolism and autoimmune thyroid health? Here he is again!
- TAGAP codes for a protein involved in T-cell signaling; like other genes in this list, is linked to autoimmune disorders such as rheumatoid arthritis, Type 1 diabetes, and multiple sclerosis.
- THEMIS-coded proteins are related to T-cell maturation, appear in lymphoid tissues, and are highly expressed in celiac disease.
These genes, and others like them, are probably involved in a wide variety of immune system function.
For instance, Type 1 diabetes and celiac disease share HLA-DQ, IL2/IL21, CCR3 and SH2B3 SNPs in populations of European ancestry. And vitamin D seems to interact with IL2RA and TAGAP.
There’s no test on this, of course.
Just get the general idea: It’s complicated.
HLA-DQ
You may remember that in Chapter 6, we talked about the human leukocyte antigen (HLA) gene complex that codes for major histocompatibility complex (MHC) proteins in humans.
These proteins, which are found on the surfaces and membranes of cells, regulate our immune system.
HLA-DQ is a type of protein that appears on the membrane of what are known as antigen-presenting cells, cells that tell other cells of the immune system (such as T cells) that there’s trouble spotted (such as a pathogen), and it’s time to go to work.
The most significant genetic risk factor for celiac disease seems to encode the HLA-DQ2/DQ8 heterodimers (molecular complexes of two macromolecules stuck together).
When immune cells that have HLA-DQ2 or DQ8 on their membranes come in contact with gluten (for instance, in the small intestine), this complex can incorrectly tell the immune system to attack the threat — in this case, healthy tissues. This creates the symptoms of celiac disease, such as gastrointestinal pain and diarrhea.
However, as we’ve seen, other genetic variants may also contribute. There are several dozen known factors so far. Some genes appear to be altered only in adults or children with celiac disease, suggesting that age may also change the genetic expression of the disease.
Not everyone with genetic variations will develop the disease, but most people who have celiac disease do seem to share some related genetic variants.
For instance, 23AndMe tests for a subtype of HLA-DQ2 called HLA-DQ2.5, using a SNP called rs2187668 located in one of the genes encoding HLA-DQ2.5.
This subtype is found in roughly 15% of the general population but in over 90% of people with celiac disease, though only about 3% of people with this specific variation will actually develop celiac disease.
23andMe also tests for rs6822844, a SNP that lies in a block of genes (KIAA1109/Tenr/IL2/IL21) that, along with another SNP in the same area (rs13119723), are strongly associated with autoimmune disease, including celiac disease.
Like many chronic diseases with a strong genetic component, the prevalence of celiac disease varies with geography and population.
Celiac disease is most often found in populations of European descent (North America, Europe, Australia, some parts of South America) as well as populations in India. It is relatively rare in most local populations in Asia.
Yet celiac disease prevalence is increasing quickly. This suggests that environmental or other physiological factors (such as the health or diversity of our gut microbiota) may be contributing.
Getting diagnostically tested for celiac disease is a painful business. In order to produce enough antibodies to show up on a lab test, patients must eat more gluten. You can imagine how that feels if you already have a sensitivity. It’s sort of like having to get stung by more bees if you have a potential bee venom allergy.
So, in this case, genetic testing is a much more pleasant option. However:
- The solution would still be the same: remove gluten from the diet.
- Research suggests that non-celiac gluten sensitivity (sensitivity to gluten without overt antibodies) is a thing.
Non-celiac gluten sensitivity
Most people in our co-author Krista’s extended family have trouble with wheat and some other grains. They get skin rashes, coughs and snuffles, even intestinal bleeding (in worst cases). You’d think that this would be an obvious case of genetically-determined celiac disease, and perhaps in some folks, it could be.
Krista, too, avoids wheat, because the connection between eating it and inflammatory symptoms is pretty clear. Yet Krista’s 23andMe test for celiac risk showed that based on a few different markers, she actually had half the average risk of celiac.
So what gives?
Is Krista’s family just full of hypochondriacs who are hating on pasta?
Or is there another explanation?
Many other frustrated people who know they don’t respond well to wheat may, nevertheless, have been told that they don’t have legitimate celiac disease, so they should quit bellyaching (so to speak).
In fact, we are coming to realize that non-celiac gluten sensitivity (NCGS) can also be a problem.
With NCGS, we don’t see the same chemical markers (such as immunoglobulin A, or IgA, antibodies), or the destruction of intestinal tissue that we see with full-blown celiac disease, but we still see inflammation.
At this point, there’s no direct-to-consumer test for NCGS, although in research labs we can look at whether inflammatory markers (such as interleukins or immune system proteins) are elevated.
For instance, the researchers in the Nutrigenomix lab looked at the relationship between genetic markers of celiac and α2-macroglobulin, a protein that goes up in response to inflammation. They found that eating more gluten was correlated with more α2-macroglobulin, but this happened regardless of people’s HLA-DQ.
That said, over half of people who have NCGS do carry similar genetic variants as people will full-blown celiac disease, and both NCGS and celiac sufferers are much more likely to have these variants than the general population.
What we found in our sample
With the HLA-DQA rs2187668 SNP tested by 23andMe, each copy of a thymine (T) increases celiac risk. So TTs (thymine-thymine) are most at risk.
We had no TTs in our sample, but about 16% of the sample had CT (cytosine-thymine), a slightly increased risk.
Several people said they were “definitely” intolerant of wheat. Four of those had had a celiac test done, which suggests that they had enough wheat intolerance symptoms to check.
Yet none of these wheat-intolerant folks were CTs. All were CCs — the lowest-risk group for celiac.
Moreover:
- All the people who said they were “definitely” intolerant to wheat were CCs, which, again, is the group that theoretically shouldn’t have problems.
- A few people said they were “definitely” not intolerant, and could crush croissants with no problems. Most, not surprisingly, were CCs, but one of those was a CT.
What this means for you
- If you suspect you may have celiac disease, see your doctor. Genetic testing can help you see if your risk of celiac disease is higher, but other tests can also confirm whether you have active antibodies to gluten.
- Research suggests that even if you do not have celiac disease, you may still have a sensitivity to gluten or other related proteins. This may be genetic or may also be related to a wide variety of other factors, such as your lifestyle, your gastrointestinal health, and your environment.
- If you notice gluten intolerance or other food sensitivities, consider working with a nutrition coach or dietitian to come up with menus that accommodate these.
Food intolerances
Lactase persistence / lactose tolerance
Lactase is an enzyme, secreted by the brush border cells of the small intestine, that helps us digest lactose, a sugar in milk.
Almost all of us can digest lactose when we are born. We need to: Breast milk is our only source of nourishment. But not all of us keep this ability as we age.
Being able to make lactase into adulthood — a trait known as lactase persistence — is determined almost completely by our genes, although other factors (such as gut microbiota) can also affect how we digest milk and dairy.
In Europeans, particularly northern and western Europeans (such as Scandinavians and Irish), the gene that codes for lactase has remained fairly common, which probably reflects the importance of milk and dairy foods for European populations. The same is true of particular farming populations in eastern / northern Africa, the Middle East, or northeast Asia where herding and dairy consumption are common.
Other populations who don’t consume as much milk and dairy — for instance, people with southeast Asian ancestry — tend not to carry this gene.
In fact, not being able to digest lactose is more the global norm than the exception. Some estimates suggest that worldwide, about 65% of adults cannot digest lactose.
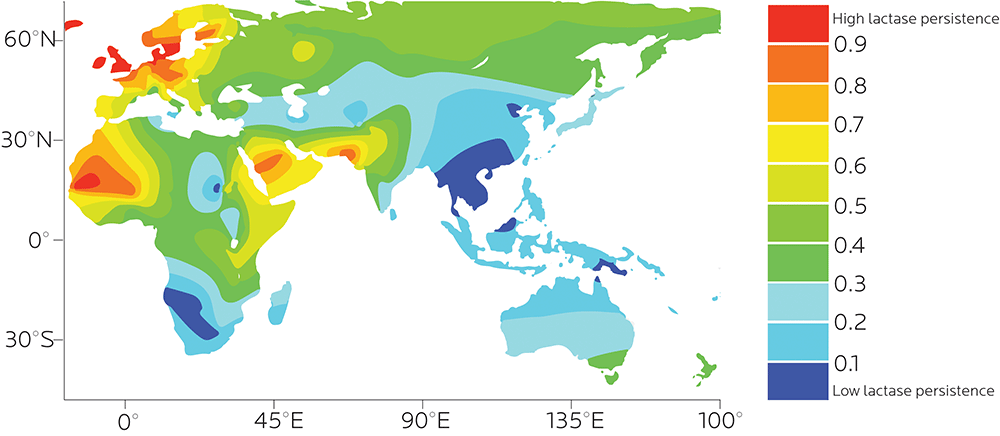
The ability to digest lactose probably evolved separately in different populations.
LCT
At the moment, we know of many different alleles of LCT, the lactose tolerance gene, that can affect this ability.
These alleles tend to travel with different haplotypes. This means that the outcome (being able to digest lactose, or not) is the same, but the reason (a specific gene variant) can be different, depending on a person’s ancestry.
For example:
- −13838G/A, −13906T/A and −13908C/T variations may have appeared independently in Tibetans, a traditionally cattle and yak-herding population.
Fun factoid!
Tibetans have also crossed domestic cattle (Bos primigenius) with yaks (Bos grunniens) to create a hybrid called a dzo.
- −13910*T (aka rs4988235) appears in Europeans, and correlates almost perfectly with lactose tolerance. This variant almost never appears in Sub-Saharan African populations, though it does appear among the Fulani of Sudan, who originally migrated from western Africa. It also appears in South America among populations with a history of European colonization.
- −13907*G, which is an East African variation appearing in Afro-Asiatic Beja populations and in the Afro-Asiatic Kenyan populations.
- −13915*G, the founder mutation for the unusually high lactase persistence in Saudi Arabians and Bedouins — which may not relate to cow’s milk but to camel’s milk. (Despite their high lactose tolerance, the characteristically European −13910*T variation doesn’t seem to appear much in this population either.)
- −14009 T>G, one of a few Ethiopian variations.
- −14010*C, typically found in a single haplotype in Kenya and Tanzania (known as Nilo-Saharan, from people who lived along the Nile and Sahara).
Other candidate variations still being tested include:
- −13779 G>C, found to date only in Amhara people (aka Abyssinians) originating in the northern and central highlands of Ethiopia.
- −13806*G, found only in Ethiopian milk drinkers, but actually associated more with non-digestion.
- −13909 C>T, another European variation.
You may remember in our previous chapter that we explored the idea of ethnic ancestry. Superficial characteristics (such as skin color) don’t actually tell us much about a person’s full set of genes.
Supporting this idea is the fact that there are at least seven different rare alleles associated with lactase persistence in one small ethnic group (Somali cattle herders from Ethiopia). In terms of understanding genetic variation, simply lumping this group into a large category called “African” or “black” would be useless at best.
MCM6 and enhancers
Along with variations upstream of the LCT gene, there is evidence that related variants in the minichromosome maintenance complex component 6 (MCM6) could also play a role.
Although MCM6 doesn’t directly affect lactose digestion, it contains two of the regulatory regions for LCT.
You’ll hopefully remember that back in Chapter 2, we talked about how genes are made, the process that regulates whether a gene is made or not, and a key region of DNA called the enhancer.
Enhancers for different genes are different, since you don’t want all the genes to be made at the same time. You want some to be turned on, and others to be turned off, depending on the situation.
If enhancers vary genetically, expression of other genes can vary too.
That’s what happens with the LCT gene and MCM6. Some people have a particular SNP (rs49882359) in the MCM6 gene that increases the activity of the LCT enhancer and leads to the expression of the LCT gene and most lactase.
Sweeps, spreads, and selection
These genetic variations, and the other possible variations that we haven’t yet found, give us clear evidence of convergent evolution, or a trait emerging independently, more than once (rather than developing in a straight linear path).
As an example of convergent evolution, consider the concept of food in “wrap” format.
For instance, wrap-type foods include sushi hand rolls, rice paper wraps, dosas, crepes, burritos, shawarma, and so forth — all having evolved independently in world cuisines. Sushi didn’t directly evolve from burritos (although now we have the sushi burrito, another one of Nature’s magnificent miracles).
In this case, we have a situation where various populations are able to produce lactase, but by different pathways.
Some of these variations also give us evidence of what is known as a selective sweep. In a selective sweep, a particular variation becomes much more common among a population, to the point where the alternative may be completely lost. Once 100% of the population have a particular form of the gene, that form is said to be “fixed”.
Selective sweeps can happen when a beneficial mutation starts out relatively rare, but quickly moves through the population, pushing out genetic rivals. This can often happen when environmental conditions change and favor the new mutation.
For example, if climate rapidly changes, genes that were previously neutral but now help the organism adapt to the new climate will likely spread.
When the Ice Age suddenly hits, everyone with the genes for a furry pelt, short limbs, and plenty of subcutaneous fat will be left standing, while everyone with the genes for squeaky-smooth-as-a-dolphin skin, lanky heat-dispersing limbs, and six-pack abs will be quickly wiped out.
Lactase persistence is an example of positive selection: a genetic mutation that promotes the emergence of new phenotypes, usually by offering some kind of advantage. (The other type of selection is purifying selection, which helps keep an existing phenotype the same.)
Many have speculated that lactase persistence emerged over and over among various populations because it was helpful for pastoral populations who might have consumed a lot of milk and dairy from cows, sheep, goats, and camels.
Genetic studies seem to support this hypothesis, since populations with higher lactase persistence also tend to include milk and dairy as part of their regular diets. (After all, try to imagine northern Europe without cheese or yogurt — no Icelandic skyr, no British Stilton, no Swiss fondue.)
Indeed, early humans don’t appear to have had much lactase persistence. The genetic variations that now let some of us enjoy ice cream and milkshakes emerged relatively recently — perhaps 10,000 to 25,000 years ago or so (some estimates even put it around 3,000 years ago).
This, again, suggests that we actively adapted to innovations in agriculture, otherwise known as “figuring out how to make animals hold still long enough for us to milk them”. Consistent dietary pressure in a small population can actually help us make pretty quick genetic changes, perhaps within 150-400 generations.
This milk-lactase persistence connection isn’t true for all populations.
For instance, Dinka and Nuer in Sudan and Somalis in Ethiopia don’t seem to have lactase persistence despite consuming dairy. In genetically lactose-intolerant populations that consume dairy but seem to digest it fine, gut microflora may also be helping out.
What we found in our sample
23andMe tests for the rs4988235 SNP of the LCT gene. The GG (guanine-guanine) variant is more likely to be lactose intolerant.
In this case, having the GG variant did seem to predict dairy intolerance. All the people who said they had significant problems digesting dairy were GGs.
That said:
- Lactose tolerance still varied. One gut-of-steel GG said they had absolutely no problems with dairy, while other GGs said their tolerance was sometimes an issue, sometimes not.
- Some AAs (adenine-adenine, the lowest-risk group) also said they had some problems digesting dairy, though nobody said it was as serious as some of the GGs did.
AG or CT?
When we were writing up our results for the rs4988235 SNP of the LCT gene, and checking both 23andMe and Nutrigenomix’s reports, we stumbled across something that seemed confusing.
- 23andMe listed the SNP variants as A/G. In other words, the 3 options would be AA, AG, or GG.
- Nutrigenomix listed them as C/T. In other words, the 3 options would be CC, CT, or TT.
Which one was right?
As it turns out, both of them.
Genetic analysis uses what are called “reference genomes”.
If you ask the question about what variant an individual has, you have to know what they vary from.
Researchers have settled on reference genomes, which are compiled and released as a set by the Genome Reference Consortium, an international consortium of experts from some of the top research institutions.
The current human reference genome is called GRCh38.p11 (GRC human genome build 38, patch level 11), released in July 2017.
(Use that little factoid to stimulate interesting conversations at cocktail parties! It is a crowd-pleaser!)
Remember that DNA has two complementary strands.
For example, if there’s an A on one strand, you know there’s a T at the same position on the opposite strand. If there’s a C on one, you know it has a G buddy.
Sometimes, when a build of a reference genome is released, the opposite strand is used.
The identifiers for SNPs are unique to each genome build, so the position might be reported as A in one build because one strand is sequenced, and a T in the next build because the complementary strand is sequenced.
For determining SNP variation, this is fine — the location varies and you don’t really care about it.
However, we discovered that 23andMe always uses the letter on the forward (i.e., transcribed) strand, independent of the reference genome.
This is an unusual choice. Most research and clinical literature use the genotypes defined in the reference genome.
You can see why 23andMe might make this choice, though — it makes it easy to say “This is the A or T that gets transcribed, so this is the one we will use”.
What this means for you
- If you can’t digest lactase, it may be genetic.
- 23andMe tests for the rs4988235 SNP of the LCT gene.
- If you still want to enjoy dairy but it gives you trouble, you have a few options.
- You can try lactase pills or lactose-free milk.
- You may find that fermented dairy (e.g. yogurt, kefir, etc.) is OK, as the process of bacterial fermentation tends to break down sugars. Supplementation with a probiotic may also help.
- If you can digest lactase, It may be from one (or more) of a number of different gene variants, depending on your ancestry. Congratulations! Enjoy the lattes, thanks to convergent evolution.
- There are other reasons you may be dairy-intolerant. For instance, your gut microbiota can play a role; you can also be sensitive to other proteins in dairy.
- If you have been diagnosed with lactose intolerance, or if you’re noticing other food sensitivities, consider working with a nutrition coach or dietitian to come up with menu options and strategies to accommodate your food needs.
Hereditary fructose intolerance
Hereditary fructose intolerance (HFI), as its name suggests, is a genetic condition in which people can’t break down fructose properly. Fructose is found in fruit, as well as many processed foods (such as soda).
People with HFI can’t make enough aldolase B, an enzyme encoded by the ALDOB gene that helps metabolize fructose. If aldolase B isn’t working properly, our bodies can’t properly convert sugar into energy, which can result in hypoglycemia (low blood sugar).
Hypoglycemia, if severe and left untreated, can lead to seizures, coma and death. In addition, the accumulation of partially metabolized fructose molecules becomes toxic to the cells and causes liver and kidney damage.
People who have HFI may avoid many sweets and fruit instinctively, which means they may never get properly diagnosed. This also means that researchers aren’t completely sure how common HFI really is.
HFI is an autosomal recessive disorder, which means that to have HFI, a person must inherit two copies of the ALDOB gene variant that causes the problem. While it can be fatal if not properly treated, people can remain healthy and free of symptoms if they avoid fructose.
There are at least 40 known ALDOB mutations that have been linked to HFI.
23andMe looks at four of the most common ALDOB mutations in people with European ancestry:
- rs1800546, aka A149P (which accounts for about 65% of all HFI-causing mutations in this population)
- rs76917243, aka A174D (accounting for about 11-14% of mutations)
- rs78340951, aka N334K (about 5-8%)
- rs387906225, aka delta4E4 (about 3%)
Together, these mutations make up about 75% of the HFI-causing mutations in this population.
What we found in our sample
Luckily for our PN team members, nobody had two of the ALDOB HFI mutations tested for: rs1800546 and rs76917243.
What this means for you
- If you’re concerned about HFI, consult your doctor. You may still have an ALDOB mutation or have HFI even if your genetic tests for these particular variations are negative.
- Even if you do not have HFI, you may find that you have digestive symptoms from eating particular types of carbohydrates known as FODMAPs: fermentable oligo-, di-, mono-saccharides and polyols. This group includes fructose.
- If you have been diagnosed with HFI, or if you’re noticing other food sensitivities, consider working with a nutrition coach or dietitian to come up with menu options and strategies to accommodate your food needs.
What’s up next
In the next chapter, we’ll look at how genetic factors other than food intolerances or sensitivities can affect how we absorb and use nutrients.





